|
Symbol Name ID |
Nrip1
nuclear receptor interacting protein 1 MGI:1315213 |
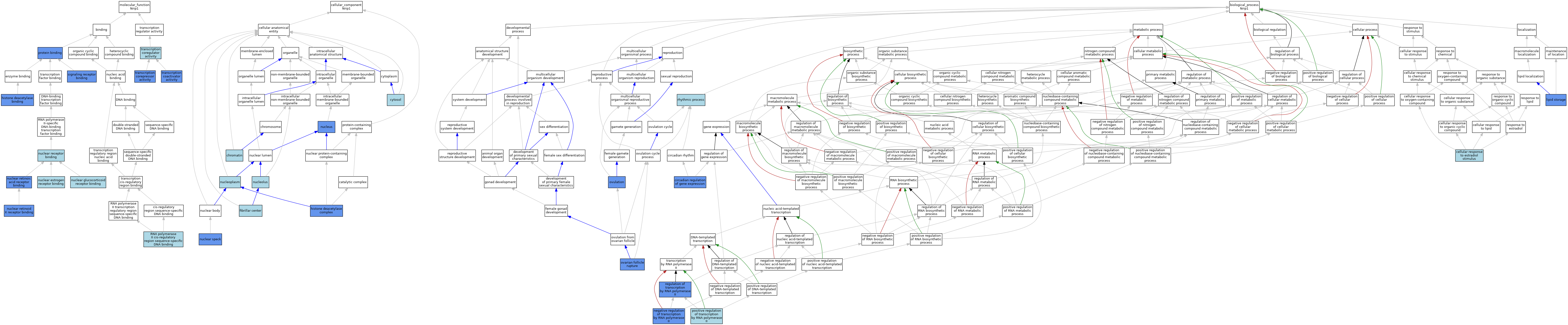
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042826 | histone deacetylase binding | IPI | J:113911 | |||||||||
| Molecular Function | GO:0030331 | nuclear estrogen receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030331 | nuclear estrogen receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0030331 | nuclear estrogen receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0035259 | nuclear glucocorticoid receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035259 | nuclear glucocorticoid receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016922 | nuclear receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042974 | nuclear retinoic acid receptor binding | IPI | J:113776 | |||||||||
| Molecular Function | GO:0046965 | nuclear retinoid X receptor binding | IPI | J:113776 | |||||||||
| Molecular Function | GO:0046965 | nuclear retinoid X receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:113911 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:175152 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245382 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:213898 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:50521 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:88316 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | IPI | J:50521 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | IDA | J:213898 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003712 | transcription coregulator activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IDA | J:114147 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IDA | J:50521 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0001650 | fibrillar center | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000118 | histone deacetylase complex | IPI | J:114064 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | IDA | J:192857 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:114719 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:154112 | |||||||||
| Biological Process | GO:0071392 | cellular response to estradiol stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0071392 | cellular response to estradiol stimulus | IBA | J:265628 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | IMP | J:213898 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | IBA | J:265628 | |||||||||
| Biological Process | GO:0019915 | lipid storage | IMP | J:90688 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:50521 | |||||||||
| Biological Process | GO:0001543 | ovarian follicle rupture | IMP | J:114726 | |||||||||
| Biological Process | GO:0030728 | ovulation | IMP | J:74971 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IDA | J:154112 | |||||||||
| Biological Process | GO:0048511 | rhythmic process | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||