|
Symbol Name ID |
Hipk1
homeodomain interacting protein kinase 1 MGI:1314873 |
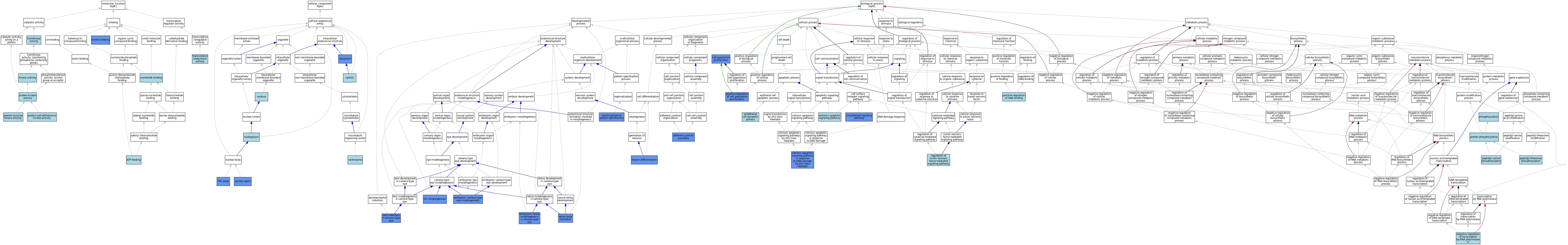
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:81640 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:65176 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:181102 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | ISS | J:50387 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004713 | protein tyrosine kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | ISS | J:50387 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005813 | centrosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:162323 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:81640 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | IDA | J:81640 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISS | J:50387 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016605 | PML body | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016605 | PML body | IDA | J:81640 | |||||||||
| Biological Process | GO:0034333 | adherens junction assembly | IMP | J:197686 | |||||||||
| Biological Process | GO:0009952 | anterior/posterior pattern specification | IGI | J:106931 | |||||||||
| Biological Process | GO:0009952 | anterior/posterior pattern specification | IGI | J:106931 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | IGI | J:106931 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | IGI | J:106931 | |||||||||
| Biological Process | GO:0048596 | embryonic camera-type eye morphogenesis | IMP | J:162323 | |||||||||
| Biological Process | GO:0060059 | embryonic retina morphogenesis in camera-type eye | IMP | J:162323 | |||||||||
| Biological Process | GO:0072577 | endothelial cell apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | IBA | J:265628 | |||||||||
| Biological Process | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | IGI | J:106931 | |||||||||
| Biological Process | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | IGI | J:106931 | |||||||||
| Biological Process | GO:0061072 | iris morphogenesis | IMP | J:162323 | |||||||||
| Biological Process | GO:0060235 | lens induction in camera-type eye | IMP | J:162323 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISS | J:50387 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IMP | J:162323 | |||||||||
| Biological Process | GO:0018105 | peptidyl-serine phosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0018107 | peptidyl-threonine phosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IGI | J:106931 | |||||||||
| Biological Process | GO:0043388 | positive regulation of DNA binding | ISS | J:50387 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | ISS | J:50387 | |||||||||
| Biological Process | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0010842 | retina layer formation | IMP | J:162323 | |||||||||
| Biological Process | GO:0007224 | smoothened signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007224 | smoothened signaling pathway | IGI | J:106931 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||