|
Symbol Name ID |
Ppp1r12a
protein phosphatase 1, regulatory subunit 12A MGI:1309528 |
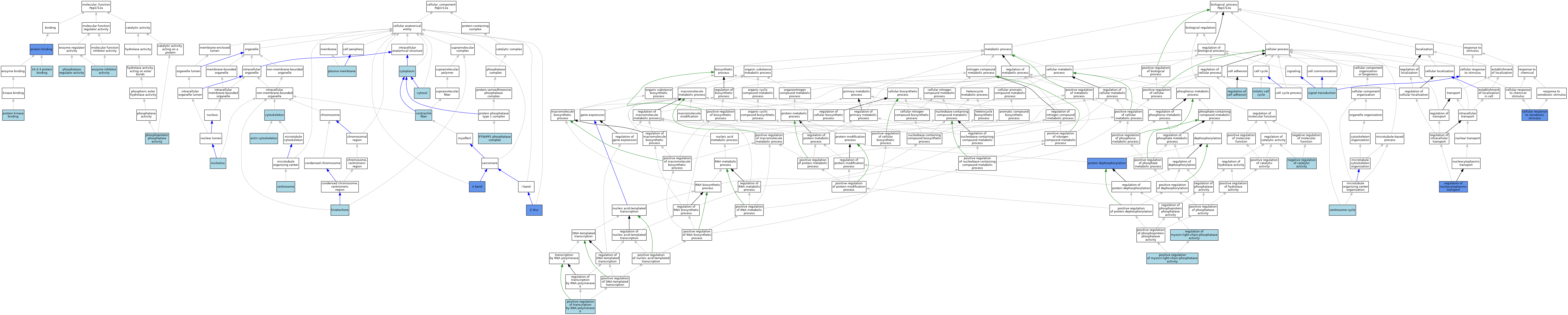
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0071889 | 14-3-3 protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004857 | enzyme inhibitor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004857 | enzyme inhibitor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019208 | phosphatase regulator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019208 | phosphatase regulator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004721 | phosphoprotein phosphatase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:166394 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:134634 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031672 | A band | IDA | J:224746 | |||||||||
| Cellular Component | GO:0031672 | A band | IBA | J:265628 | |||||||||
| Cellular Component | GO:0015629 | actin cytoskeleton | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005813 | centrosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043292 | contractile fiber | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0072357 | PTW/PP1 phosphatase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030018 | Z disc | IDA | J:224746 | |||||||||
| Cellular Component | GO:0030018 | Z disc | IBA | J:265628 | |||||||||
| Biological Process | GO:0071466 | cellular response to xenobiotic stimulus | IDA | J:138635 | |||||||||
| Biological Process | GO:0007098 | centrosome cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0000278 | mitotic cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0043086 | negative regulation of catalytic activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0006470 | protein dephosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006470 | protein dephosphorylation | IMP | J:119479 | |||||||||
| Biological Process | GO:0030155 | regulation of cell adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0035507 | regulation of myosin-light-chain-phosphatase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0046822 | regulation of nucleocytoplasmic transport | IMP | J:119479 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||