|
Symbol Name ID |
Cyp2j5
cytochrome P450, family 2, subfamily j, polypeptide 5 MGI:1270149 |
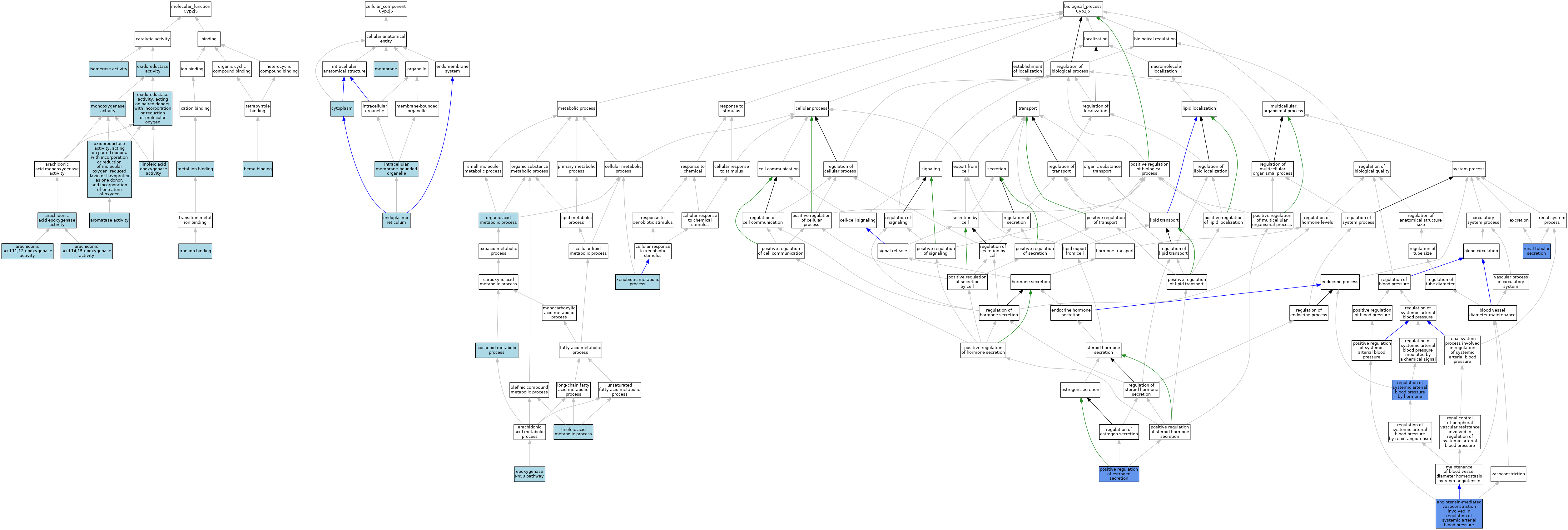
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008405 | arachidonic acid 11,12-epoxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008404 | arachidonic acid 14,15-epoxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008392 | arachidonic acid epoxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0070330 | aromatase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0020037 | heme binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005506 | iron ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016853 | isomerase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0071614 | linoleic acid epoxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004497 | monooxygenase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004497 | monooxygenase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0001998 | angiotensin-mediated vasoconstriction involved in regulation of systemic arterial blood pressure | IMP | J:146047 | |||||||||
| Biological Process | GO:0019373 | epoxygenase P450 pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0006690 | icosanoid metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043651 | linoleic acid metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006082 | organic acid metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:2000863 | positive regulation of estrogen secretion | IMP | J:146047 | |||||||||
| Biological Process | GO:0001990 | regulation of systemic arterial blood pressure by hormone | IMP | J:146047 | |||||||||
| Biological Process | GO:0097254 | renal tubular secretion | IMP | J:146047 | |||||||||
| Biological Process | GO:0006805 | xenobiotic metabolic process | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||