|
Symbol Name ID |
Arhgap6
Rho GTPase activating protein 6 MGI:1196332 |
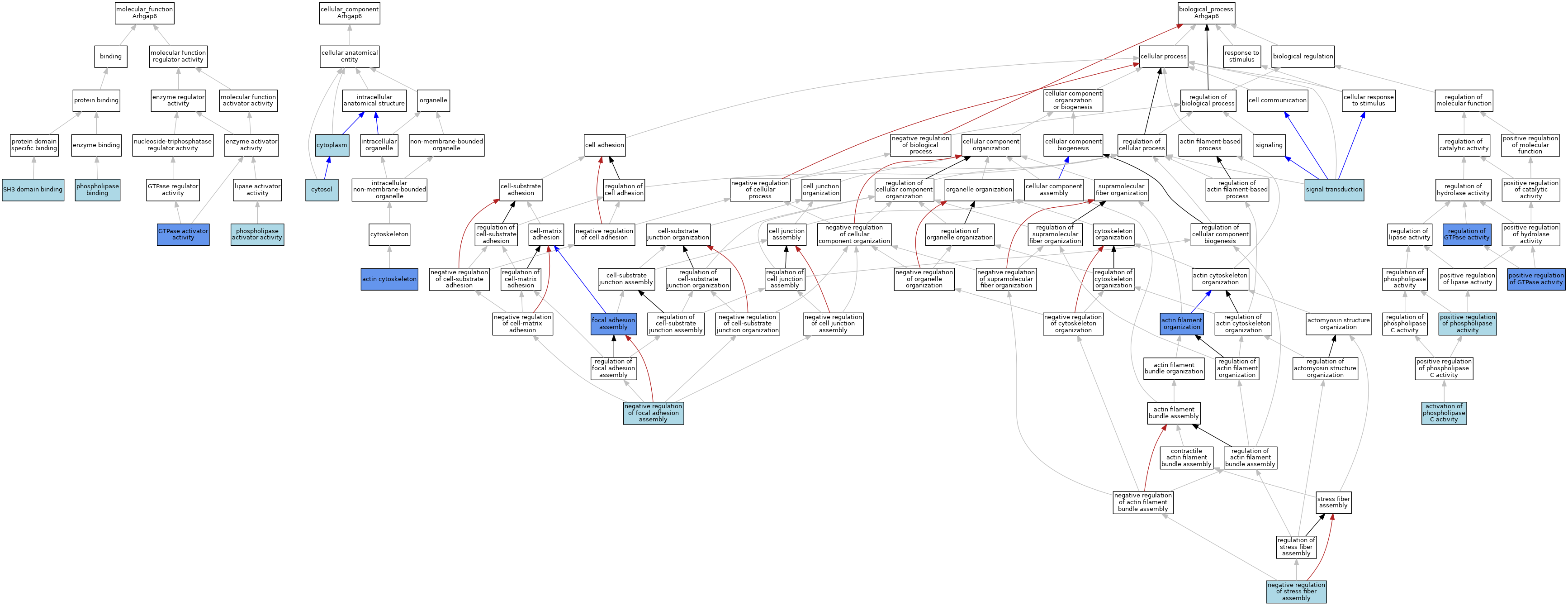
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005096 | GTPase activator activity | IDA | J:80946 | |||||||||
| Molecular Function | GO:0016004 | phospholipase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016004 | phospholipase activator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043274 | phospholipase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043274 | phospholipase binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | IEA | J:60000 | |||||||||
| Cellular Component | GO:0015629 | actin cytoskeleton | IDA | J:80946 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Biological Process | GO:0007015 | actin filament organization | IDA | J:80946 | |||||||||
| Biological Process | GO:0007202 | activation of phospholipase C activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0007202 | activation of phospholipase C activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0048041 | focal adhesion assembly | IDA | J:80946 | |||||||||
| Biological Process | GO:0051895 | negative regulation of focal adhesion assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0051497 | negative regulation of stress fiber assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0043547 | positive regulation of GTPase activity | IDA | J:80946 | |||||||||
| Biological Process | GO:0010518 | positive regulation of phospholipase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0043087 | regulation of GTPase activity | IDA | J:80946 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||