|
Symbol Name ID |
Kl
klotho MGI:1101771 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
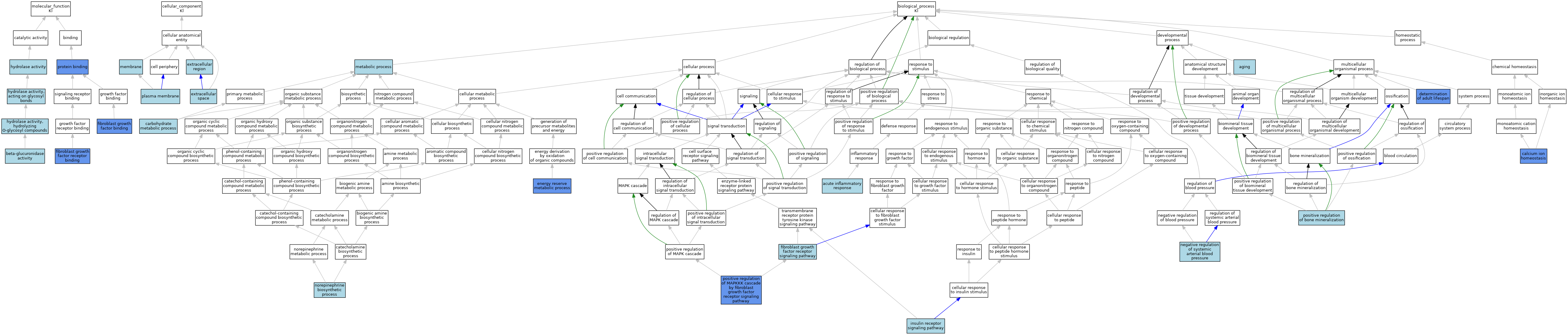
| Molecular Function | GO:0004566 | beta-glucuronidase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0017134 | fibroblast growth factor binding | IPI | J:117479 | |||||||||
| Molecular Function | GO:0017134 | fibroblast growth factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0017134 | fibroblast growth factor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005104 | fibroblast growth factor receptor binding | IPI | J:117479 | |||||||||
| Molecular Function | GO:0005104 | fibroblast growth factor receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016798 | hydrolase activity, acting on glycosyl bonds | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:123536 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:156501 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:117479 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:110570 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:110570 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:110570 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | ISS | J:46369 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISS | J:46369 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0002526 | acute inflammatory response | ISO | J:155856 | |||||||||
| Biological Process | GO:0007568 | aging | ISO | J:164563 | |||||||||
| Biological Process | GO:0055074 | calcium ion homeostasis | IGI | J:183281 | |||||||||
| Biological Process | GO:0005975 | carbohydrate metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0008340 | determination of adult lifespan | IMP | J:44109 | |||||||||
| Biological Process | GO:0006112 | energy reserve metabolic process | IMP | J:66078 | |||||||||
| Biological Process | GO:0006112 | energy reserve metabolic process | IMP | J:66078 | |||||||||
| Biological Process | GO:0008543 | fibroblast growth factor receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0008286 | insulin receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0008152 | metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0003085 | negative regulation of systemic arterial blood pressure | ISO | J:155856 | |||||||||
| Biological Process | GO:0042421 | norepinephrine biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0030501 | positive regulation of bone mineralization | ISO | J:164563 | |||||||||
| Biological Process | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway | IGI | J:110570 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||