|
Symbol Name ID |
Atxn3
ataxin 3 MGI:1099442 |
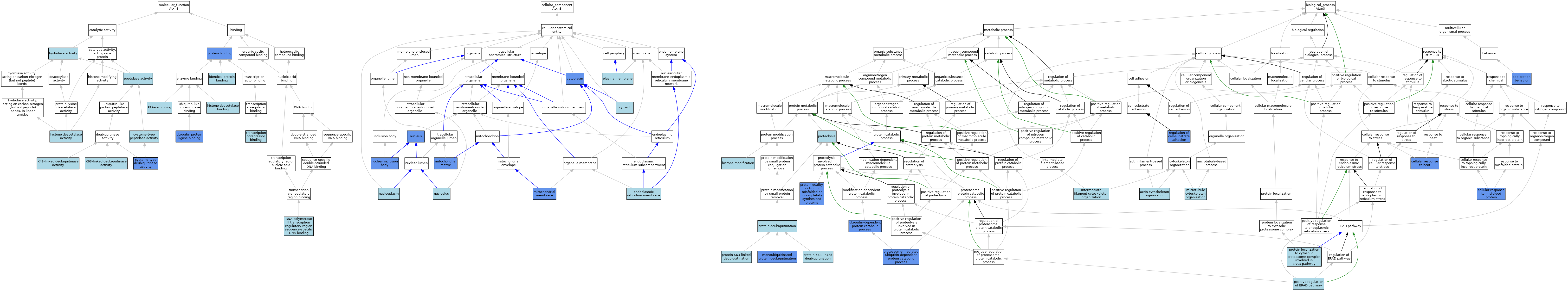
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0051117 | ATPase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004843 | cysteine-type deubiquitinase activity | IDA | J:183193 | |||||||||
| Molecular Function | GO:0004843 | cysteine-type deubiquitinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008234 | cysteine-type peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004407 | histone deacetylase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042826 | histone deacetylase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990380 | K48-linked deubiquitinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0061578 | K63-linked deubiquitinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:145209 | |||||||||
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0001222 | transcription corepressor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | IPI | J:183193 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:136856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:155356 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:122993 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:165379 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005759 | mitochondrial matrix | IDA | J:136856 | |||||||||
| Cellular Component | GO:0031966 | mitochondrial membrane | IDA | J:136856 | |||||||||
| Cellular Component | GO:0042405 | nuclear inclusion body | IDA | J:122993 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:136856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:165379 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:155356 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | ISO | J:73065 | |||||||||
| Biological Process | GO:0034605 | cellular response to heat | IMP | J:155356 | |||||||||
| Biological Process | GO:0071218 | cellular response to misfolded protein | IMP | J:183193 | |||||||||
| Biological Process | GO:0035640 | exploration behavior | IMP | J:145048 | |||||||||
| Biological Process | GO:0016570 | histone modification | ISO | J:155856 | |||||||||
| Biological Process | GO:0045104 | intermediate filament cytoskeleton organization | ISO | J:73065 | |||||||||
| Biological Process | GO:0000226 | microtubule cytoskeleton organization | ISO | J:73065 | |||||||||
| Biological Process | GO:0035520 | monoubiquitinated protein deubiquitination | IDA | J:183193 | |||||||||
| Biological Process | GO:1904294 | positive regulation of ERAD pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | IMP | J:183193 | |||||||||
| Biological Process | GO:0016579 | protein deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0016579 | protein deubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0071108 | protein K48-linked deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0070536 | protein K63-linked deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:1904379 | protein localization to cytosolic proteasome complex involved in ERAD pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | IMP | J:183193 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0010810 | regulation of cell-substrate adhesion | IMP | J:165449 | |||||||||
| Biological Process | GO:0010810 | regulation of cell-substrate adhesion | ISO | J:73065 | |||||||||
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | IMP | J:145048 | |||||||||
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | IMP | J:145048 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||