|
Symbol Name ID |
Ifnar2
interferon (alpha and beta) receptor 2 MGI:1098243 |
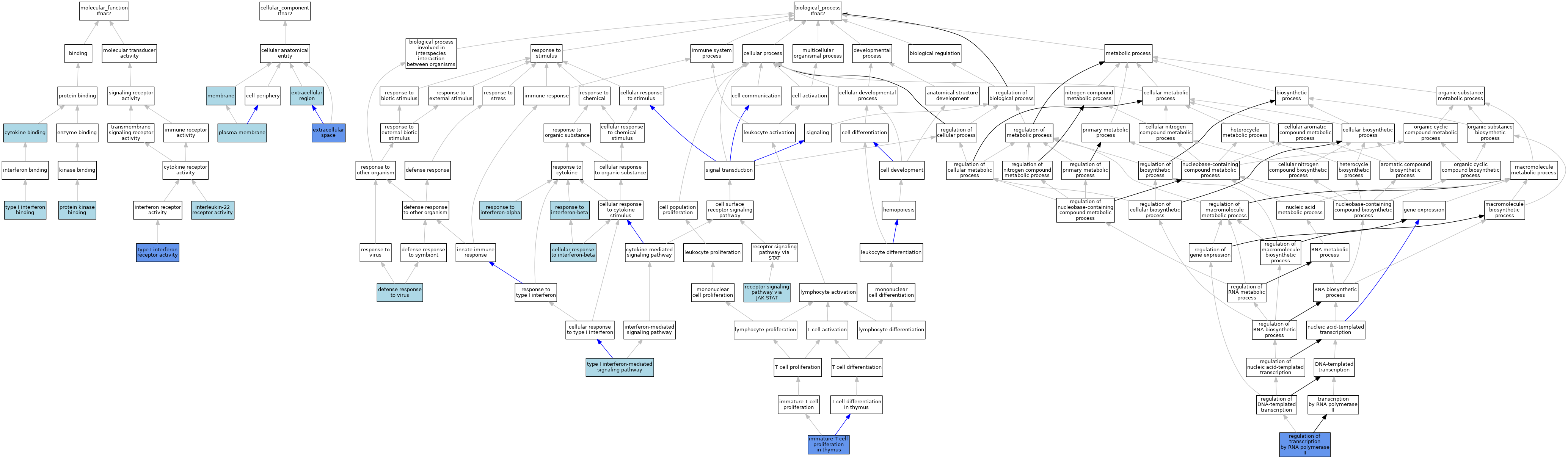
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019955 | cytokine binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042018 | interleukin-22 receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019962 | type I interferon binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004905 | type I interferon receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004905 | type I interferon receptor activity | IDA | J:66964 | |||||||||
| Molecular Function | GO:0004905 | type I interferon receptor activity | IDA | J:43022 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | HDA | J:221550 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:66964 | |||||||||
| Cellular Component | GO:0016020 | membrane | TAS | J:43022 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0035458 | cellular response to interferon-beta | ISO | J:164563 | |||||||||
| Biological Process | GO:0051607 | defense response to virus | ISO | J:164563 | |||||||||
| Biological Process | GO:0033080 | immature T cell proliferation in thymus | IDA | J:66964 | |||||||||
| Biological Process | GO:0007259 | receptor signaling pathway via JAK-STAT | ISO | J:164563 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IDA | J:42947 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IDA | J:66964 | |||||||||
| Biological Process | GO:0035455 | response to interferon-alpha | ISO | J:164563 | |||||||||
| Biological Process | GO:0035456 | response to interferon-beta | ISO | J:164563 | |||||||||
| Biological Process | GO:0060337 | type I interferon-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0060337 | type I interferon-mediated signaling pathway | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||