|
Symbol Name ID |
P2rx1
purinergic receptor P2X, ligand-gated ion channel, 1 MGI:1098235 |
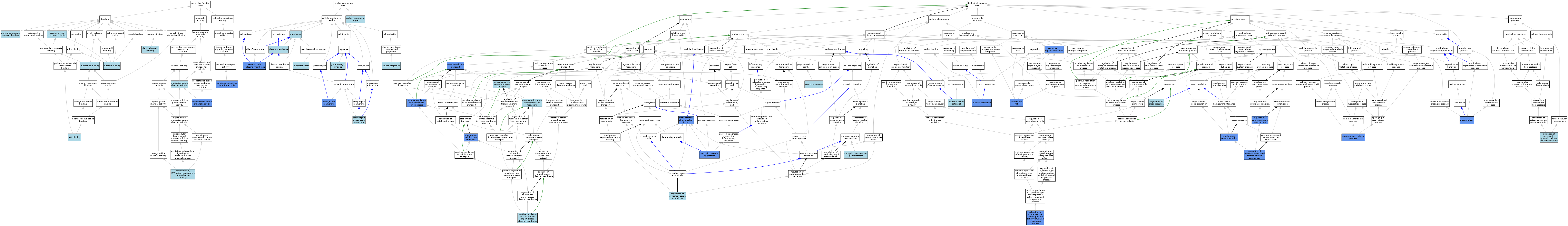
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004931 | extracellularly ATP-gated monoatomic cation channel activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004931 | extracellularly ATP-gated monoatomic cation channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005261 | monoatomic cation channel activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0005261 | monoatomic cation channel activity | IMP | J:59336 | |||||||||
| Molecular Function | GO:0005216 | monoatomic ion channel activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0097159 | organic cyclic compound binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0001614 | purinergic nucleotide receptor activity | IMP | J:59336 | |||||||||
| Molecular Function | GO:0001614 | purinergic nucleotide receptor activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0043924 | suramin binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IDA | J:102580 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | IDA | J:74184 | |||||||||
| Cellular Component | GO:0048787 | presynaptic active zone membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Biological Process | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | IDA | J:110330 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0046513 | ceramide biosynthetic process | IDA | J:110330 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IMP | J:59336 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IGI | J:118491 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IMP | J:118491 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IGI | J:118491 | |||||||||
| Biological Process | GO:0007320 | insemination | IMP | J:59336 | |||||||||
| Biological Process | GO:0098655 | monoatomic cation transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0034220 | monoatomic ion transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IGI | J:118491 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IMP | J:118491 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IGI | J:118491 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IGI | J:118491 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | ISO | J:73065 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IMP | J:118491 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IGI | J:118491 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IMP | J:59336 | |||||||||
| Biological Process | GO:0019228 | neuronal action potential | ISO | J:155856 | |||||||||
| Biological Process | GO:0030168 | platelet activation | IMP | J:85102 | |||||||||
| Biological Process | GO:1905665 | positive regulation of calcium ion import across plasma membrane | ISO | J:155856 | |||||||||
| Biological Process | GO:0043270 | positive regulation of monoatomic ion transport | IGI | J:118491 | |||||||||
| Biological Process | GO:0043270 | positive regulation of monoatomic ion transport | IGI | J:118491 | |||||||||
| Biological Process | GO:0043270 | positive regulation of monoatomic ion transport | IMP | J:118491 | |||||||||
| Biological Process | GO:0008217 | regulation of blood pressure | ISO | J:155856 | |||||||||
| Biological Process | GO:0051924 | regulation of calcium ion transport | IMP | J:116400 | |||||||||
| Biological Process | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration | ISO | J:155856 | |||||||||
| Biological Process | GO:0006940 | regulation of smooth muscle contraction | IMP | J:59336 | |||||||||
| Biological Process | GO:0006940 | regulation of smooth muscle contraction | IMP | J:102580 | |||||||||
| Biological Process | GO:2000300 | regulation of synaptic vesicle exocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0003056 | regulation of vascular associated smooth muscle contraction | IMP | J:116400 | |||||||||
| Biological Process | GO:0019229 | regulation of vasoconstriction | IMP | J:118486 | |||||||||
| Biological Process | GO:0033198 | response to ATP | IMP | J:118486 | |||||||||
| Biological Process | GO:0033198 | response to ATP | IMP | J:101793 | |||||||||
| Biological Process | GO:0010033 | response to organic substance | IMP | J:118491 | |||||||||
| Biological Process | GO:0002554 | serotonin secretion by platelet | IMP | J:85102 | |||||||||
| Biological Process | GO:0035249 | synaptic transmission, glutamatergic | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||