|
Symbol Name ID |
S1pr1
sphingosine-1-phosphate receptor 1 MGI:1096355 |
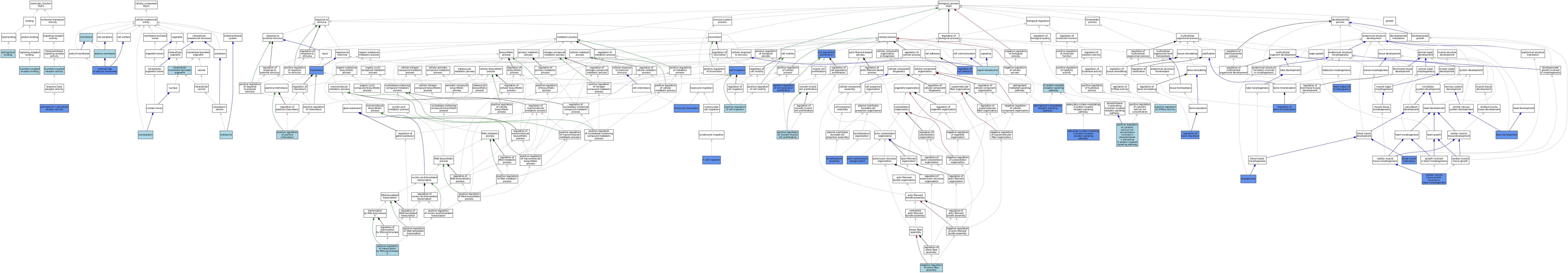
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0001664 | G protein-coupled receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046625 | sphingolipid binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0038036 | sphingosine-1-phosphate receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0038036 | sphingosine-1-phosphate receptor activity | IMP | J:71961 | |||||||||
| Molecular Function | GO:0038036 | sphingosine-1-phosphate receptor activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IMP | J:141431 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0031532 | actin cytoskeleton reorganization | IMP | J:120048 | |||||||||
| Biological Process | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | IDA | J:71372 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IDA | J:93030 | |||||||||
| Biological Process | GO:0001955 | blood vessel maturation | IMP | J:71961 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:103755 | |||||||||
| Biological Process | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis | IMP | J:173601 | |||||||||
| Biological Process | GO:0016477 | cell migration | IMP | J:147106 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | IMP | J:103755 | |||||||||
| Biological Process | GO:0006935 | chemotaxis | IMP | J:71961 | |||||||||
| Biological Process | GO:0006935 | chemotaxis | IMP | J:120048 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0061384 | heart trabecula morphogenesis | IMP | J:173601 | |||||||||
| Biological Process | GO:0030032 | lamellipodium assembly | IMP | J:120048 | |||||||||
| Biological Process | GO:0030595 | leukocyte chemotaxis | IDA | J:219661 | |||||||||
| Biological Process | GO:0051497 | negative regulation of stress fiber assembly | ISO | J:155856 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:155856 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IMP | J:103755 | |||||||||
| Biological Process | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0043547 | positive regulation of GTPase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0050927 | positive regulation of positive chemotaxis | ISO | J:155856 | |||||||||
| Biological Process | GO:0048661 | positive regulation of smooth muscle cell proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:155856 | |||||||||
| Biological Process | GO:0030500 | regulation of bone mineralization | IMP | J:147106 | |||||||||
| Biological Process | GO:0045124 | regulation of bone resorption | IMP | J:147106 | |||||||||
| Biological Process | GO:0030155 | regulation of cell adhesion | IDA | J:93030 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | IMP | J:71961 | |||||||||
| Biological Process | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0072678 | T cell migration | IMP | J:89522 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||