|
Symbol Name ID |
Taf1b
TATA-box binding protein associated factor, RNA polymerase I, B MGI:109577 |
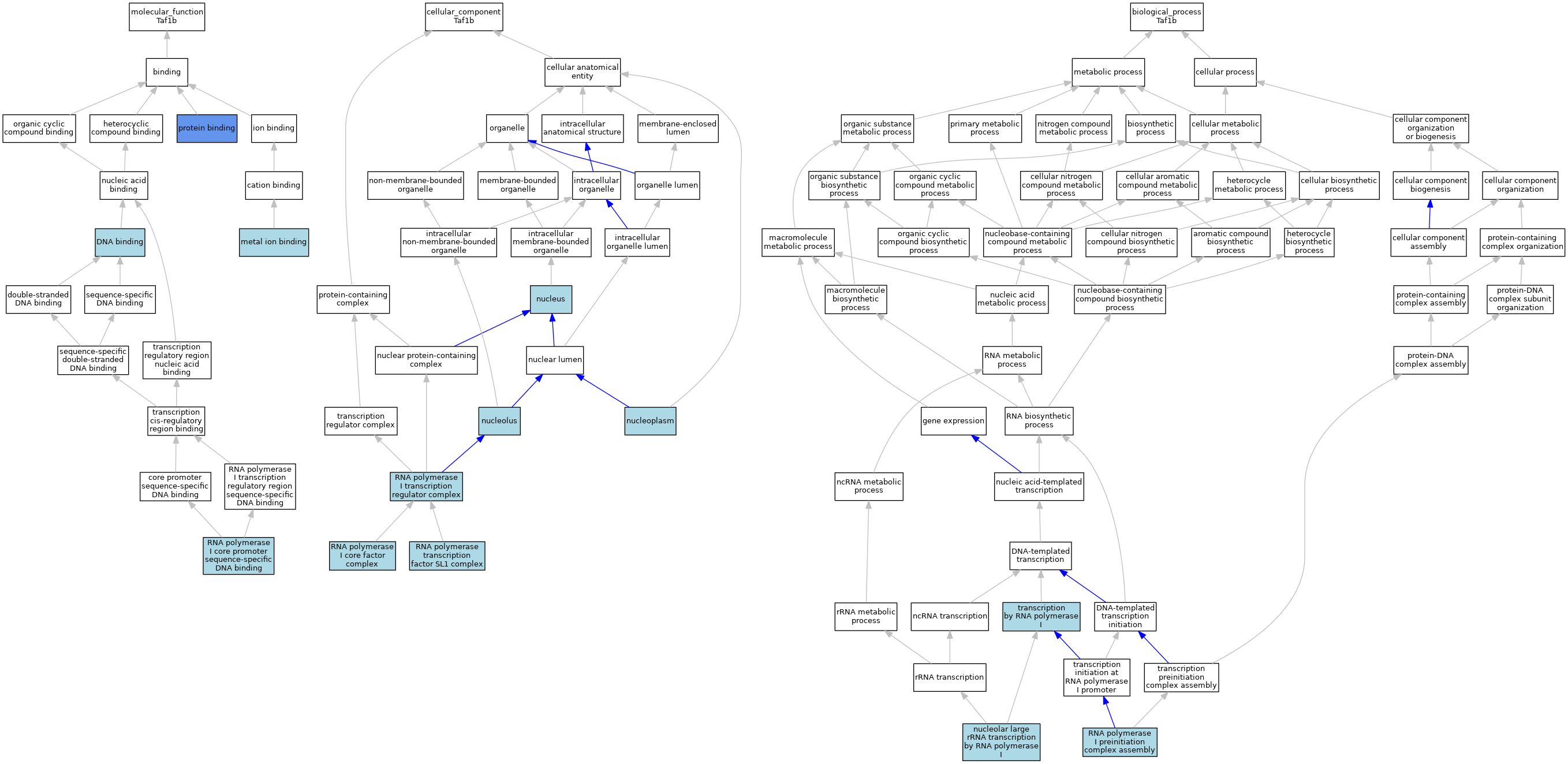
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:173136 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:39012 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:39012 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:39012 | |||||||||
| Molecular Function | GO:0001164 | RNA polymerase I core promoter sequence-specific DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001164 | RNA polymerase I core promoter sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5211233 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5211235 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0070860 | RNA polymerase I core factor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0070860 | RNA polymerase I core factor complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000120 | RNA polymerase I transcription regulator complex | TAS | J:39012 | |||||||||
| Cellular Component | GO:0005668 | RNA polymerase transcription factor SL1 complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | IBA | J:265628 | |||||||||
| Biological Process | GO:0001188 | RNA polymerase I preinitiation complex assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0006360 | transcription by RNA polymerase I | TAS | J:39012 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||