|
Symbol Name ID |
Ctsc
cathepsin C MGI:109553 |
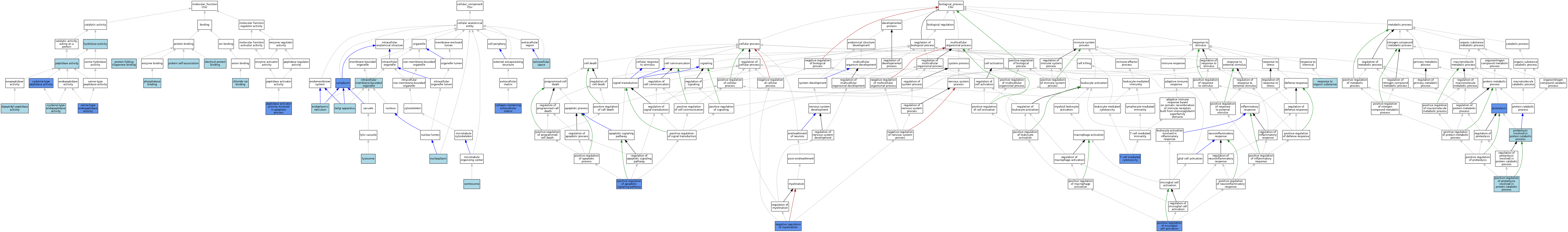
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0031404 | chloride ion binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004197 | cysteine-type endopeptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008234 | cysteine-type peptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008234 | cysteine-type peptidase activity | IDA | J:242069 | |||||||||
| Molecular Function | GO:0008239 | dipeptidyl-peptidase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016505 | peptidase activator activity involved in apoptotic process | IGI | J:165930 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0019902 | phosphatase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0043621 | protein self-association | ISO | J:155856 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | IMP | J:165930 | |||||||||
| Cellular Component | GO:0005813 | centrosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | HDA | J:265607 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:242069 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Biological Process | GO:0031642 | negative regulation of myelination | IDA | J:242069 | |||||||||
| Biological Process | GO:0031642 | negative regulation of myelination | IMP | J:242069 | |||||||||
| Biological Process | GO:2001235 | positive regulation of apoptotic signaling pathway | IGI | J:165930 | |||||||||
| Biological Process | GO:1903980 | positive regulation of microglial cell activation | IDA | J:242069 | |||||||||
| Biological Process | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006508 | proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IMP | J:165930 | |||||||||
| Biological Process | GO:0051603 | proteolysis involved in protein catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0010033 | response to organic substance | ISO | J:155856 | |||||||||
| Biological Process | GO:0001913 | T cell mediated cytotoxicity | IGI | J:165930 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||