|
Symbol Name ID |
Pdcd6
programmed cell death 6 MGI:109283 |
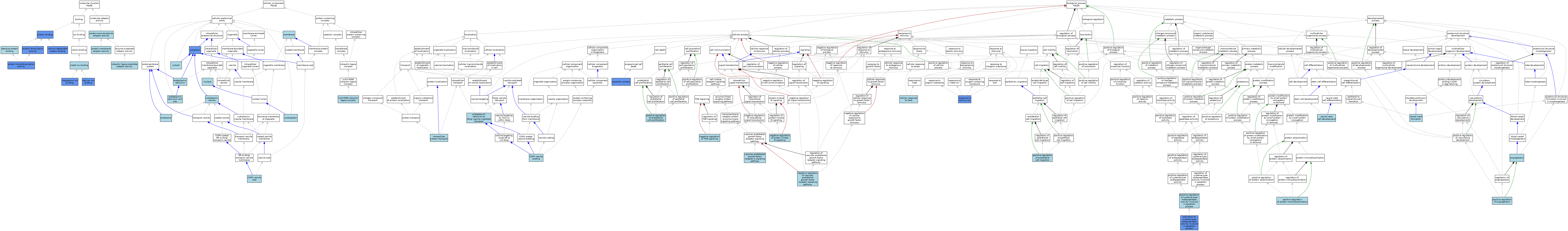
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | IMP | J:127172 | |||||||||
| Molecular Function | GO:0048306 | calcium-dependent protein binding | IPI | J:127172 | |||||||||
| Molecular Function | GO:0048306 | calcium-dependent protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000287 | magnesium ion binding | IDA | J:240322 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:201254 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:177167 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:161035 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:87334 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:87334 | |||||||||
| Molecular Function | GO:0046983 | protein dimerization activity | IPI | J:53664 | |||||||||
| Molecular Function | GO:0046983 | protein dimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | IPI | J:53664 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | IDA | J:240322 | |||||||||
| Molecular Function | GO:0030674 | protein-macromolecule adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043495 | protein-membrane adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990756 | ubiquitin ligase-substrate adaptor activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030127 | COPII vesicle coat | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031463 | Cul3-RING ubiquitin ligase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:127172 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0070971 | endoplasmic reticulum exit site | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | IDA | J:87334 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IDA | J:33763 | |||||||||
| Biological Process | GO:0034605 | cellular response to heat | ISO | J:164563 | |||||||||
| Biological Process | GO:0048208 | COPII vesicle coating | ISO | J:164563 | |||||||||
| Biological Process | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006886 | intracellular protein transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0051898 | negative regulation of protein kinase B signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0032007 | negative regulation of TOR signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0014032 | neural crest cell development | ISO | J:164563 | |||||||||
| Biological Process | GO:0014029 | neural crest formation | ISO | J:164563 | |||||||||
| Biological Process | GO:0045766 | positive regulation of angiogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0010595 | positive regulation of endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0001938 | positive regulation of endothelial cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:1902527 | positive regulation of protein monoubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0051592 | response to calcium ion | IDA | J:127172 | |||||||||
| Biological Process | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||