|
Symbol Name ID |
Npas2
neuronal PAS domain protein 2 MGI:109232 |
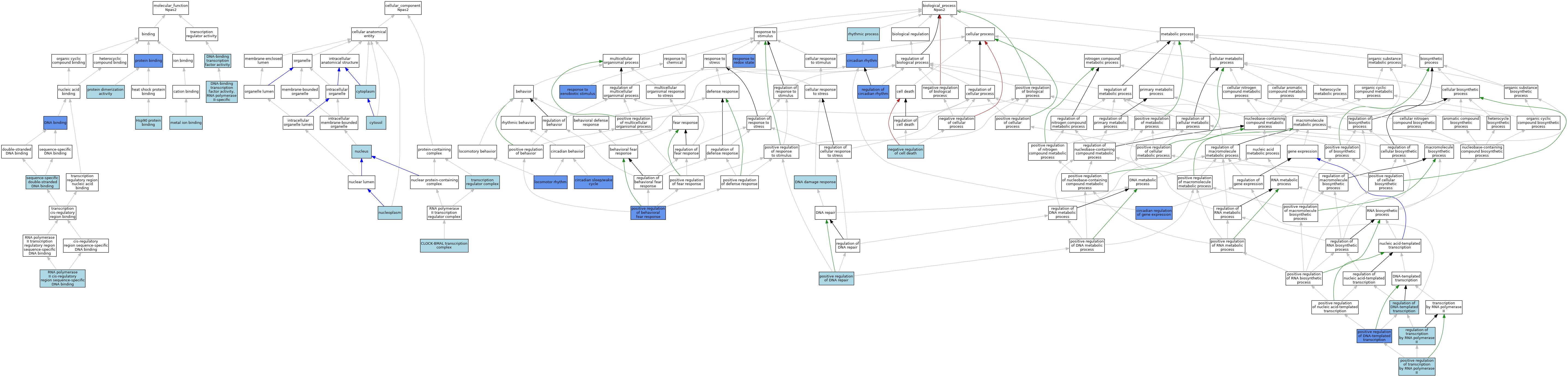
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IDA | J:209471 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0051879 | Hsp90 protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:207820 | |||||||||
| Molecular Function | GO:0046983 | protein dimerization activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:1990513 | CLOCK-BMAL transcription complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-508619 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-508656 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-549451 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5663263 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-508656 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-508741 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663114 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663129 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663146 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663147 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663158 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663161 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663169 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663172 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663185 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5663189 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5669295 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-8878686 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005667 | transcription regulator complex | ISO | J:73065 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | IMP | J:135812 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | IMP | J:202325 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | IMP | J:271703 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | IBA | J:265628 | |||||||||
| Biological Process | GO:0007623 | circadian rhythm | IEP | J:207820 | |||||||||
| Biological Process | GO:0042745 | circadian sleep/wake cycle | IMP | J:84471 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | ISO | J:164563 | |||||||||
| Biological Process | GO:0045475 | locomotor rhythm | IMP | J:84471 | |||||||||
| Biological Process | GO:0060548 | negative regulation of cell death | ISO | J:164563 | |||||||||
| Biological Process | GO:2000987 | positive regulation of behavioral fear response | IMP | J:271703 | |||||||||
| Biological Process | GO:0045739 | positive regulation of DNA repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IMP | J:274080 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IDA | J:202325 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IDA | J:135812 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:73065 | |||||||||
| Biological Process | GO:0042752 | regulation of circadian rhythm | IMP | J:202325 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0051775 | response to redox state | IDA | J:209471 | |||||||||
| Biological Process | GO:0051775 | response to redox state | ISO | J:164563 | |||||||||
| Biological Process | GO:0009410 | response to xenobiotic stimulus | IMP | J:271703 | |||||||||
| Biological Process | GO:0048511 | rhythmic process | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||