|
Symbol Name ID |
Tradd
TNFRSF1A-associated via death domain MGI:109200 |
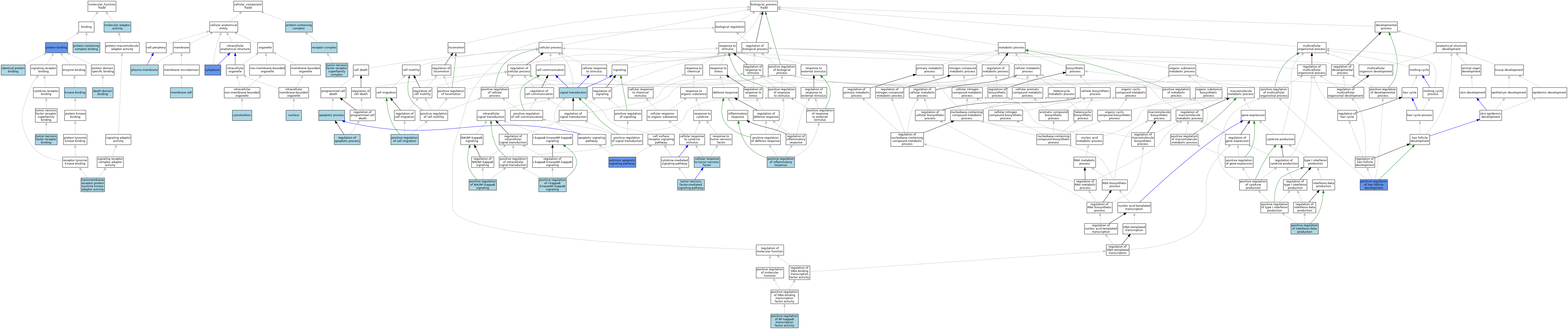
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0070513 | death domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019900 | kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0060090 | molecular adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:108710 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:281201 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:228302 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:138824 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:151999 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005164 | tumor necrosis factor receptor binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:108710 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0002947 | tumor necrosis factor receptor superfamily complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0002947 | tumor necrosis factor receptor superfamily complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | RCA | J:84168 | |||||||||
| Biological Process | GO:0071356 | cellular response to tumor necrosis factor | ISO | J:164563 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | IGI | J:202809 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | IGI | J:202809 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0051798 | positive regulation of hair follicle development | IMP | J:108710 | |||||||||
| Biological Process | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | ISO | J:280922 | |||||||||
| Biological Process | GO:0050729 | positive regulation of inflammatory response | ISO | J:164563 | |||||||||
| Biological Process | GO:0032728 | positive regulation of interferon-beta production | ISO | J:280922 | |||||||||
| Biological Process | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | ISO | J:164563 | |||||||||
| Biological Process | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0042981 | regulation of apoptotic process | RCA | J:84168 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
| Biological Process | GO:0033209 | tumor necrosis factor-mediated signaling pathway | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||