|
Symbol Name ID |
Ext2
exostosin glycosyltransferase 2 MGI:108050 |
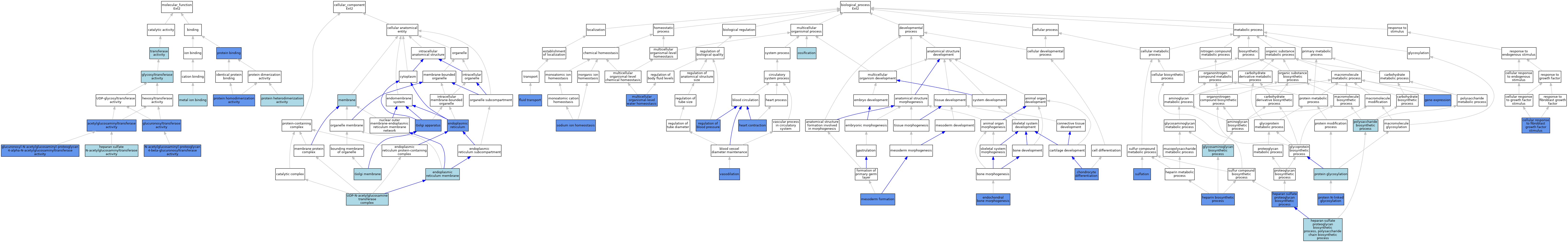
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008375 | acetylglucosaminyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008375 | acetylglucosaminyltransferase activity | IDA | J:113870 | |||||||||
| Molecular Function | GO:0008375 | acetylglucosaminyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity | IC | J:113870 | |||||||||
| Molecular Function | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity | IGI | J:113870 | |||||||||
| Molecular Function | GO:0015020 | glucuronosyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015020 | glucuronosyltransferase activity | IDA | J:113870 | |||||||||
| Molecular Function | GO:0016757 | glycosyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity | IC | J:113870 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity | IC | J:113870 | |||||||||
| Molecular Function | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity | IGI | J:113870 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:133353 | |||||||||
| Molecular Function | GO:0046982 | protein heterodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | IDA | J:113870 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:113870 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IC | J:113870 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IDA | J:113870 | |||||||||
| Cellular Component | GO:0000139 | Golgi membrane | IC | J:113870 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043541 | UDP-N-acetylglucosamine transferase complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0044344 | cellular response to fibroblast growth factor stimulus | IMP | J:286741 | |||||||||
| Biological Process | GO:0002062 | chondrocyte differentiation | IMP | J:103125 | |||||||||
| Biological Process | GO:0060350 | endochondral bone morphogenesis | IGI | J:286741 | |||||||||
| Biological Process | GO:0042044 | fluid transport | IGI | J:279714 | |||||||||
| Biological Process | GO:0010467 | gene expression | IMP | J:133353 | |||||||||
| Biological Process | GO:0010467 | gene expression | IDA | J:133353 | |||||||||
| Biological Process | GO:0006024 | glycosaminoglycan biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0060047 | heart contraction | IGI | J:279714 | |||||||||
| Biological Process | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | IGI | J:113870 | |||||||||
| Biological Process | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | IMP | J:103125 | |||||||||
| Biological Process | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | IMP | J:103125 | |||||||||
| Biological Process | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0030210 | heparin biosynthetic process | IGI | J:286741 | |||||||||
| Biological Process | GO:0001707 | mesoderm formation | IMP | J:103125 | |||||||||
| Biological Process | GO:0001707 | mesoderm formation | IMP | J:103125 | |||||||||
| Biological Process | GO:0050891 | multicellular organismal-level water homeostasis | IGI | J:279714 | |||||||||
| Biological Process | GO:0001503 | ossification | ISO | J:164563 | |||||||||
| Biological Process | GO:0000271 | polysaccharide biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006486 | protein glycosylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0006487 | protein N-linked glycosylation | IDA | J:133353 | |||||||||
| Biological Process | GO:0008217 | regulation of blood pressure | IGI | J:279714 | |||||||||
| Biological Process | GO:0055078 | sodium ion homeostasis | IGI | J:279714 | |||||||||
| Biological Process | GO:0051923 | sulfation | IDA | J:133353 | |||||||||
| Biological Process | GO:0042311 | vasodilation | IGI | J:279714 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||