|
Symbol Name ID |
Pdcd4
programmed cell death 4 MGI:107490 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
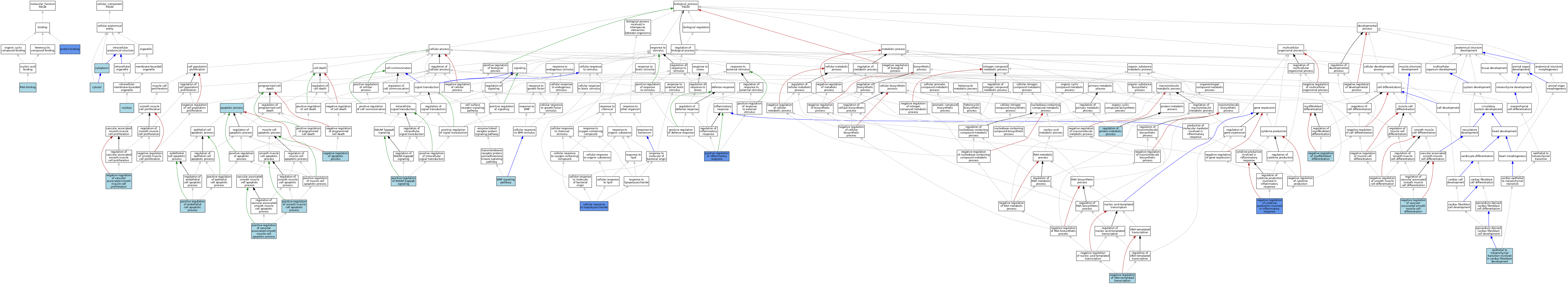
| Molecular Function | GO:0005515 | protein binding | IPI | J:145850 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:113966 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114111 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0030509 | BMP signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0071222 | cellular response to lipopolysaccharide | IMP | J:158464 | |||||||||
| Biological Process | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development | ISO | J:155856 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:1900016 | negative regulation of cytokine production involved in inflammatory response | IMP | J:158464 | |||||||||
| Biological Process | GO:1900016 | negative regulation of cytokine production involved in inflammatory response | IMP | J:120583 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:1904761 | negative regulation of myofibroblast differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:2000353 | positive regulation of endothelial cell apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0050729 | positive regulation of inflammatory response | IMP | J:158464 | |||||||||
| Biological Process | GO:0050729 | positive regulation of inflammatory response | IMP | J:120583 | |||||||||
| Biological Process | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0034393 | positive regulation of smooth muscle cell apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:1905461 | positive regulation of vascular associated smooth muscle cell apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0051246 | regulation of protein metabolic process | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||