|
Symbol Name ID |
Dnajc3
DnaJ heat shock protein family (Hsp40) member C3 MGI:107373 |
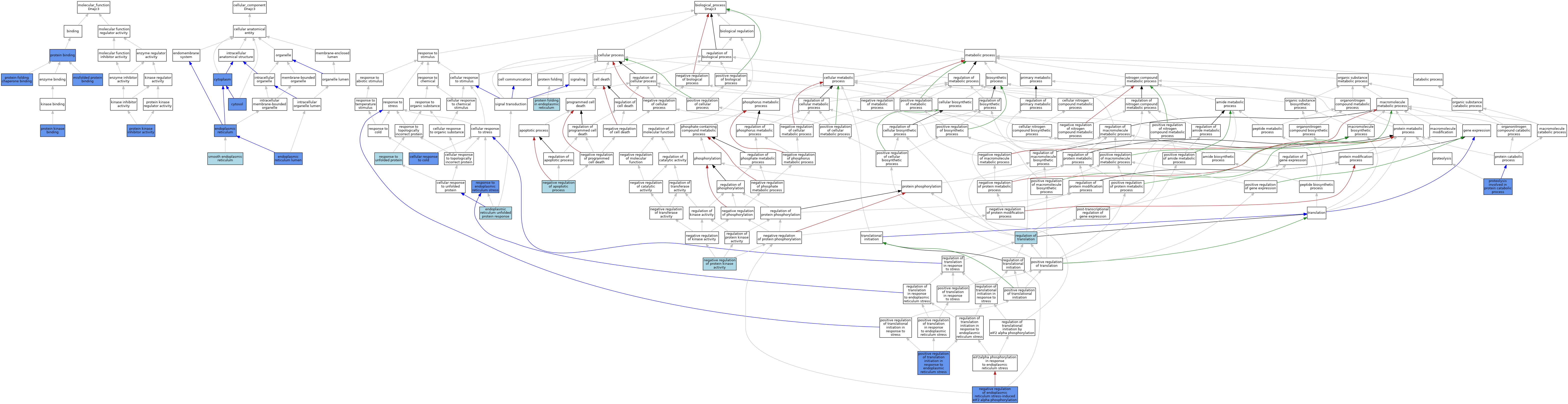
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0051787 | misfolded protein binding | IDA | J:156044 | |||||||||
| Molecular Function | GO:0051787 | misfolded protein binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:156044 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IPI | J:218404 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IDA | J:218404 | |||||||||
| Molecular Function | GO:0004860 | protein kinase inhibitor activity | IDA | J:218404 | |||||||||
| Molecular Function | GO:0004860 | protein kinase inhibitor activity | ISS | J:33042 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | IDA | J:115988 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | IPI | J:156044 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:33042 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:218404 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | J:156044 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:218404 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005788 | endoplasmic reticulum lumen | IDA | J:156044 | |||||||||
| Cellular Component | GO:0005790 | smooth endoplasmic reticulum | ISO | J:155856 | |||||||||
| Biological Process | GO:0070417 | cellular response to cold | IDA | J:218404 | |||||||||
| Biological Process | GO:0030968 | endoplasmic reticulum unfolded protein response | IC | J:156044 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | IDA | J:218404 | |||||||||
| Biological Process | GO:0006469 | negative regulation of protein kinase activity | ISS | J:33042 | |||||||||
| Biological Process | GO:0036494 | positive regulation of translation initiation in response to endoplasmic reticulum stress | IDA | J:218404 | |||||||||
| Biological Process | GO:0034975 | protein folding in endoplasmic reticulum | IBA | J:265628 | |||||||||
| Biological Process | GO:0051603 | proteolysis involved in protein catabolic process | IDA | J:156044 | |||||||||
| Biological Process | GO:0006417 | regulation of translation | IEA | J:60000 | |||||||||
| Biological Process | GO:0034976 | response to endoplasmic reticulum stress | IMP | J:115988 | |||||||||
| Biological Process | GO:0006986 | response to unfolded protein | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||