|
Symbol Name ID |
Atp2a1
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 MGI:105058 |
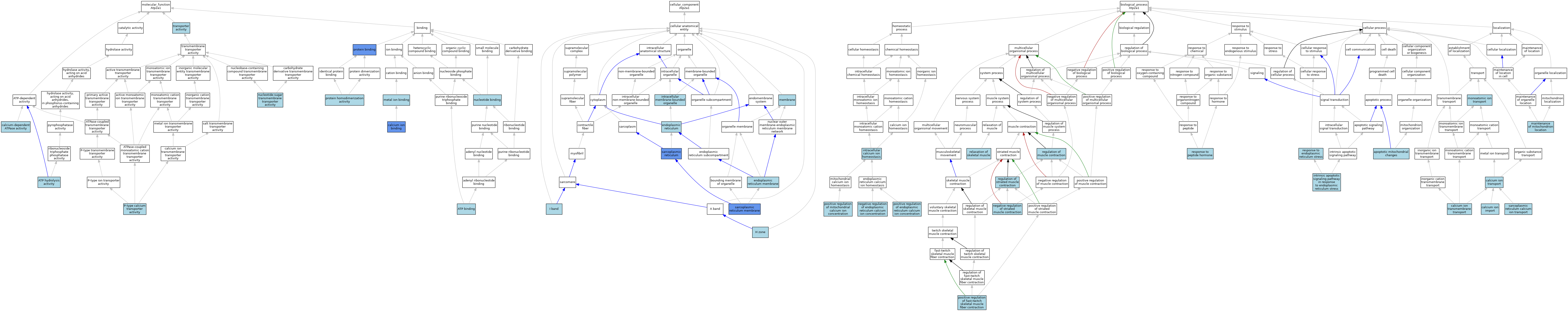
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | IDA | J:260257 | |||||||||
| Molecular Function | GO:0030899 | calcium-dependent ATPase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005338 | nucleotide-sugar transmembrane transporter activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005388 | P-type calcium transporter activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005388 | P-type calcium transporter activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005388 | P-type calcium transporter activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005388 | P-type calcium transporter activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005388 | P-type calcium transporter activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:274777 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:229305 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:217579 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005215 | transporter activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031673 | H zone | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031674 | I band | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | IDA | J:72636 | |||||||||
| Cellular Component | GO:0033017 | sarcoplasmic reticulum membrane | IDA | J:229305 | |||||||||
| Cellular Component | GO:0033017 | sarcoplasmic reticulum membrane | IDA | J:217579 | |||||||||
| Biological Process | GO:0008637 | apoptotic mitochondrial changes | ISO | J:164563 | |||||||||
| Biological Process | GO:0070509 | calcium ion import | ISO | J:164563 | |||||||||
| Biological Process | GO:0070588 | calcium ion transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0070588 | calcium ion transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0006816 | calcium ion transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006816 | calcium ion transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006816 | calcium ion transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0006816 | calcium ion transport | ISO | J:73065 | |||||||||
| Biological Process | GO:0006874 | intracellular calcium ion homeostasis | IBA | J:265628 | |||||||||
| Biological Process | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | ISO | J:164563 | |||||||||
| Biological Process | GO:0051659 | maintenance of mitochondrion location | ISO | J:164563 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | ISO | J:164563 | |||||||||
| Biological Process | GO:0045988 | negative regulation of striated muscle contraction | ISO | J:164563 | |||||||||
| Biological Process | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | ISO | J:164563 | |||||||||
| Biological Process | GO:0031448 | positive regulation of fast-twitch skeletal muscle fiber contraction | ISO | J:164563 | |||||||||
| Biological Process | GO:0051561 | positive regulation of mitochondrial calcium ion concentration | ISO | J:164563 | |||||||||
| Biological Process | GO:0006937 | regulation of muscle contraction | TAS | J:72636 | |||||||||
| Biological Process | GO:0006942 | regulation of striated muscle contraction | ISO | J:164563 | |||||||||
| Biological Process | GO:0090076 | relaxation of skeletal muscle | ISO | J:164563 | |||||||||
| Biological Process | GO:0034976 | response to endoplasmic reticulum stress | ISO | J:164563 | |||||||||
| Biological Process | GO:0043434 | response to peptide hormone | ISO | J:155856 | |||||||||
| Biological Process | GO:0070296 | sarcoplasmic reticulum calcium ion transport | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||