|
Symbol Name ID |
Ksr1
kinase suppressor of ras 1 MGI:105051 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
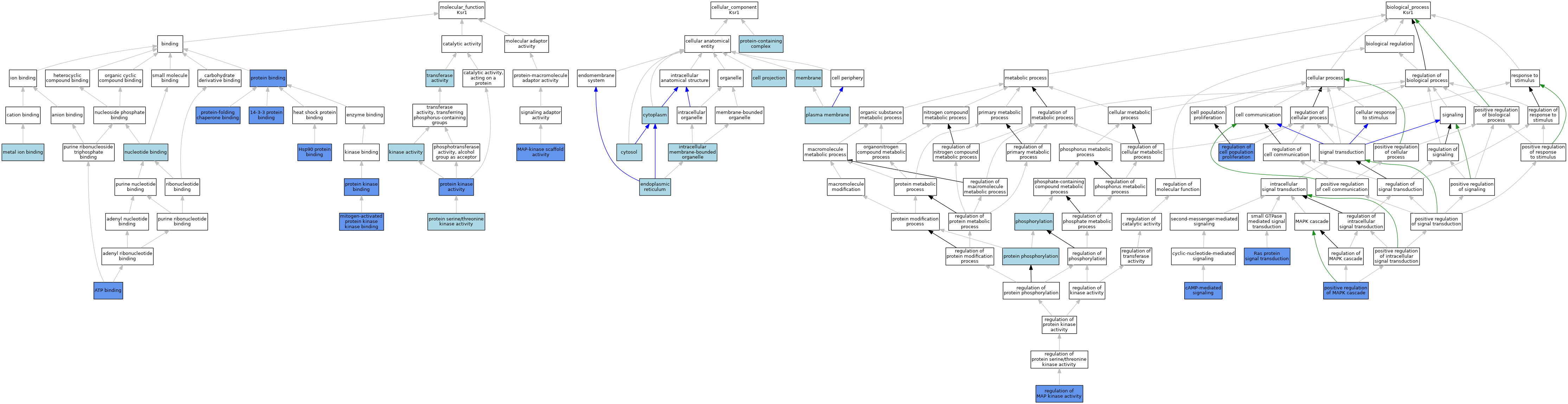
| Molecular Function | GO:0071889 | 14-3-3 protein binding | IPI | J:230400 | |||||||||
| Molecular Function | GO:0071889 | 14-3-3 protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IMP | J:277858 | |||||||||
| Molecular Function | GO:0051879 | Hsp90 protein binding | IPI | J:230400 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005078 | MAP-kinase scaffold activity | IMP | J:231274 | |||||||||
| Molecular Function | GO:0005078 | MAP-kinase scaffold activity | IMP | J:230400 | |||||||||
| Molecular Function | GO:0005078 | MAP-kinase scaffold activity | IDA | J:150410 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0031434 | mitogen-activated protein kinase kinase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0031434 | mitogen-activated protein kinase kinase binding | IPI | J:230400 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:326246 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:209645 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:184683 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:192645 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:127178 | |||||||||
| Molecular Function | GO:0005515 | protein binding | ISO | J:187529 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IMP | J:277858 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IPI | J:277858 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | IPI | J:230400 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:187529 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:187529 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:187529 | |||||||||
| Biological Process | GO:0019933 | cAMP-mediated signaling | IMP | J:231274 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IMP | J:277858 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IMP | J:230400 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IMP | J:231274 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IDA | J:150410 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0007265 | Ras protein signal transduction | IMP | J:231274 | |||||||||
| Biological Process | GO:0007265 | Ras protein signal transduction | IBA | J:265628 | |||||||||
| Biological Process | GO:0042127 | regulation of cell population proliferation | IMP | J:84644 | |||||||||
| Biological Process | GO:0043405 | regulation of MAP kinase activity | IMP | J:84644 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||