|
Symbol Name ID |
Ang2
angiogenin, ribonuclease A family, member 2 MGI:104984 |
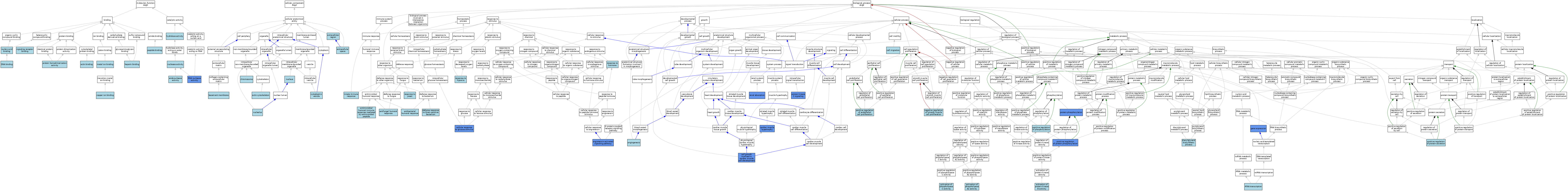
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005507 | copper ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004519 | endonuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008201 | heparin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004518 | nuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0042277 | peptide binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004540 | RNA nuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004540 | RNA nuclease activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004540 | RNA nuclease activity | IDA | J:208392 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0015629 | actin cytoskeleton | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005604 | basement membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005694 | chromosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0032431 | activation of phospholipase A2 activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0007202 | activation of phospholipase C activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0032148 | activation of protein kinase B activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0038166 | angiotensin-activated signaling pathway | IDA | J:216684 | |||||||||
| Biological Process | GO:0019731 | antibacterial humoral response | ISO | J:164563 | |||||||||
| Biological Process | GO:0019731 | antibacterial humoral response | IBA | J:265628 | |||||||||
| Biological Process | GO:0019732 | antifungal humoral response | ISO | J:164563 | |||||||||
| Biological Process | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | ISO | J:164563 | |||||||||
| Biological Process | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | IBA | J:265628 | |||||||||
| Biological Process | GO:0003300 | cardiac muscle hypertrophy | IDA | J:251673 | |||||||||
| Biological Process | GO:0061049 | cell growth involved in cardiac muscle cell development | IDA | J:251673 | |||||||||
| Biological Process | GO:0016477 | cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0071333 | cellular response to glucose stimulus | IDA | J:126723 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | ISO | J:164563 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | IBA | J:265628 | |||||||||
| Biological Process | GO:0006651 | diacylglycerol biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0010467 | gene expression | IDA | J:240742 | |||||||||
| Biological Process | GO:0010467 | gene expression | IDA | J:216684 | |||||||||
| Biological Process | GO:0045087 | innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0048662 | negative regulation of smooth muscle cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0001938 | positive regulation of endothelial cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0042327 | positive regulation of phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | IDA | J:251673 | |||||||||
| Biological Process | GO:0050714 | positive regulation of protein secretion | ISO | J:164563 | |||||||||
| Biological Process | GO:0070528 | protein kinase C signaling | IDA | J:216684 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IMP | J:216684 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IDA | J:240742 | |||||||||
| Biological Process | GO:0070293 | renal absorption | IDA | J:216684 | |||||||||
| Biological Process | GO:0009725 | response to hormone | ISO | J:164563 | |||||||||
| Biological Process | GO:0001666 | response to hypoxia | ISO | J:164563 | |||||||||
| Biological Process | GO:0001878 | response to yeast | ISO | J:164563 | |||||||||
| Biological Process | GO:0009303 | rRNA transcription | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||