|
Symbol Name ID |
Fnta
farnesyltransferase, CAAX box, alpha MGI:104683 |
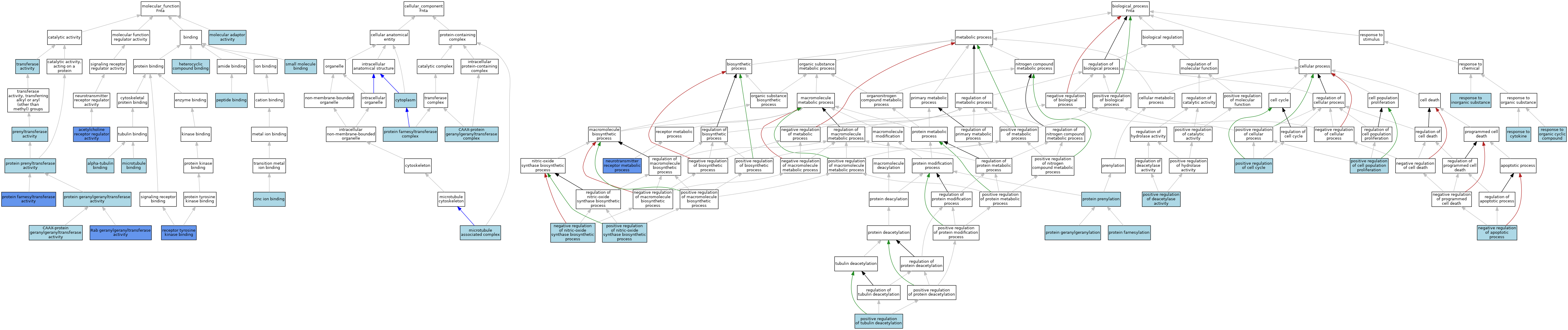
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0030548 | acetylcholine receptor regulator activity | IMP | J:86639 | |||||||||
| Molecular Function | GO:0043014 | alpha-tubulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004662 | CAAX-protein geranylgeranyltransferase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004662 | CAAX-protein geranylgeranyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:1901363 | heterocyclic compound binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0060090 | molecular adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042277 | peptide binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004659 | prenyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004660 | protein farnesyltransferase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004660 | protein farnesyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004660 | protein farnesyltransferase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004660 | protein farnesyltransferase activity | IDA | J:86639 | |||||||||
| Molecular Function | GO:0004660 | protein farnesyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004660 | protein farnesyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004661 | protein geranylgeranyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008318 | protein prenyltransferase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004663 | Rab geranylgeranyltransferase activity | IDA | J:86639 | |||||||||
| Molecular Function | GO:0030971 | receptor tyrosine kinase binding | IPI | J:86639 | |||||||||
| Molecular Function | GO:0036094 | small molecule binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005953 | CAAX-protein geranylgeranyltransferase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005953 | CAAX-protein geranylgeranyltransferase complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005953 | CAAX-protein geranylgeranyltransferase complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005875 | microtubule associated complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005965 | protein farnesyltransferase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005965 | protein farnesyltransferase complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0045213 | neurotransmitter receptor metabolic process | IMP | J:86639 | |||||||||
| Biological Process | GO:0045787 | positive regulation of cell cycle | ISO | J:155856 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0090045 | positive regulation of deacetylase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0090044 | positive regulation of tubulin deacetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0018343 | protein farnesylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0018343 | protein farnesylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0018344 | protein geranylgeranylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0018344 | protein geranylgeranylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0018342 | protein prenylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0034097 | response to cytokine | ISO | J:155856 | |||||||||
| Biological Process | GO:0010035 | response to inorganic substance | ISO | J:155856 | |||||||||
| Biological Process | GO:0014070 | response to organic cyclic compound | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||