|
Symbol Name ID |
Rps6ka1
ribosomal protein S6 kinase polypeptide 1 MGI:104558 |
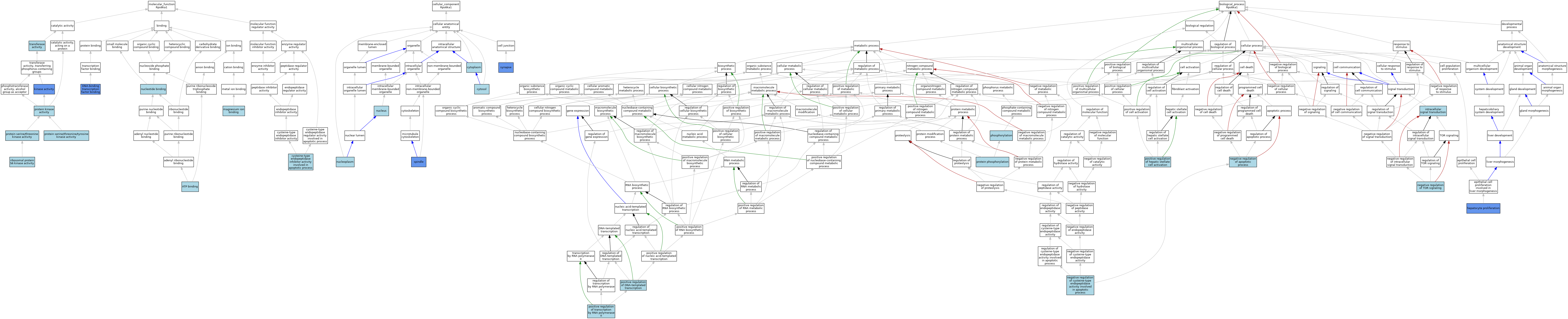
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process | ISO | J:164563 | |||||||||
| Molecular Function | GO:0140297 | DNA-binding transcription factor binding | IPI | J:133506 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IDA | J:133506 | |||||||||
| Molecular Function | GO:0000287 | magnesium ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004712 | protein serine/threonine/tyrosine kinase activity | ISO | J:199676 | |||||||||
| Molecular Function | GO:0004711 | ribosomal protein S6 kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005819 | spindle | IDA | J:93217 | |||||||||
| Cellular Component | GO:0045202 | synapse | EXP | J:263419 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:263419 | |||||||||
| Biological Process | GO:0072574 | hepatocyte proliferation | IDA | J:133506 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | IEA | J:72247 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0032007 | negative regulation of TOR signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:155856 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IBA | J:265628 | |||||||||
| Biological Process | GO:2000491 | positive regulation of hepatic stellate cell activation | ISO | J:164563 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | ISO | J:155856 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||