|
Symbol Name ID |
Relb
avian reticuloendotheliosis viral (v-rel) oncogene related B MGI:103289 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
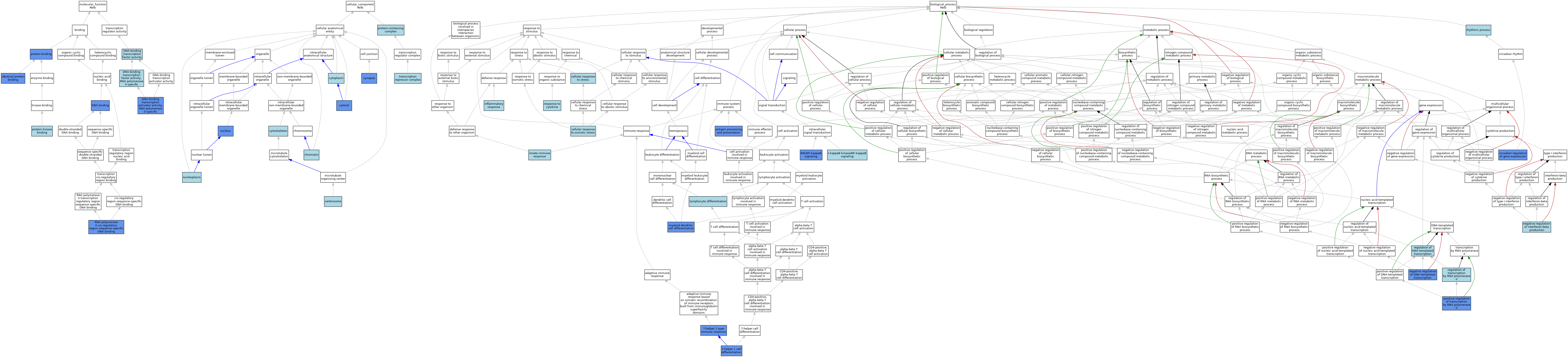
| Molecular Function | GO:0003677 | DNA binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IDA | J:100877 | |||||||||
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | IMP | J:24211 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:247386 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:211236 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:190685 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:160776 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:75807 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:75807 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IMP | J:24211 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IDA | J:211236 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005813 | centrosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:75807 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:176455 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:75807 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:184438 | |||||||||
| Cellular Component | GO:0045202 | synapse | IEP | J:184438 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:184438 | |||||||||
| Cellular Component | GO:0017053 | transcription repressor complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0019882 | antigen processing and presentation | IMP | J:23081 | |||||||||
| Biological Process | GO:0071470 | cellular response to osmotic stress | ISO | J:155856 | |||||||||
| Biological Process | GO:0033554 | cellular response to stress | IBA | J:265628 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | IMP | J:211236 | |||||||||
| Biological Process | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IBA | J:265628 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0030098 | lymphocyte differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043011 | myeloid dendritic cell differentiation | IMP | J:85808 | |||||||||
| Biological Process | GO:0043011 | myeloid dendritic cell differentiation | IMP | J:23081 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | IDA | J:211236 | |||||||||
| Biological Process | GO:0032688 | negative regulation of interferon-beta production | ISO | J:164563 | |||||||||
| Biological Process | GO:0038061 | NIK/NF-kappaB signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0038061 | NIK/NF-kappaB signaling | IDA | J:196136 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:24211 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0034097 | response to cytokine | IBA | J:265628 | |||||||||
| Biological Process | GO:0048511 | rhythmic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0045063 | T-helper 1 cell differentiation | IMP | J:107516 | |||||||||
| Biological Process | GO:0042088 | T-helper 1 type immune response | IMP | J:23081 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||