|
Symbol Name ID |
Ptprm
protein tyrosine phosphatase receptor type M MGI:102694 |
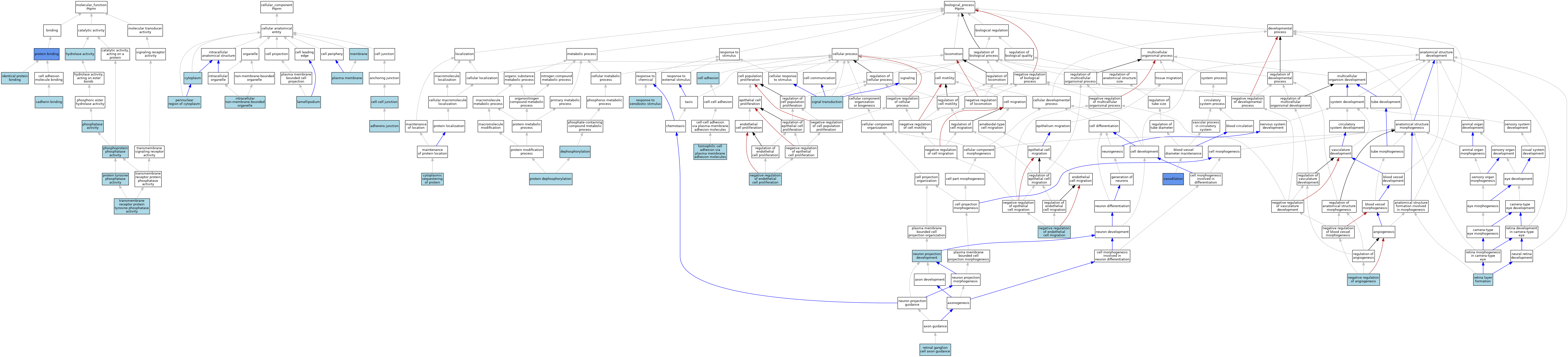
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0045296 | cadherin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016791 | phosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004721 | phosphoprotein phosphatase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:273548 | |||||||||
| Molecular Function | GO:0004725 | protein tyrosine phosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043232 | intracellular non-membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | ISO | J:97057 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IEA | J:60000 | |||||||||
| Biological Process | GO:0051220 | cytoplasmic sequestering of protein | ISO | J:164563 | |||||||||
| Biological Process | GO:0016311 | dephosphorylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | ISO | J:164563 | |||||||||
| Biological Process | GO:0016525 | negative regulation of angiogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0010596 | negative regulation of endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0001937 | negative regulation of endothelial cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0031175 | neuron projection development | ISO | J:164563 | |||||||||
| Biological Process | GO:0006470 | protein dephosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006470 | protein dephosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0009410 | response to xenobiotic stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0010842 | retina layer formation | ISO | J:164563 | |||||||||
| Biological Process | GO:0031290 | retinal ganglion cell axon guidance | ISO | J:164563 | |||||||||
| Biological Process | GO:0007165 | signal transduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0042311 | vasodilation | IMP | J:97057 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||