|
Symbol Name ID |
Scnn1a
sodium channel, nonvoltage-gated 1 alpha MGI:101782 |
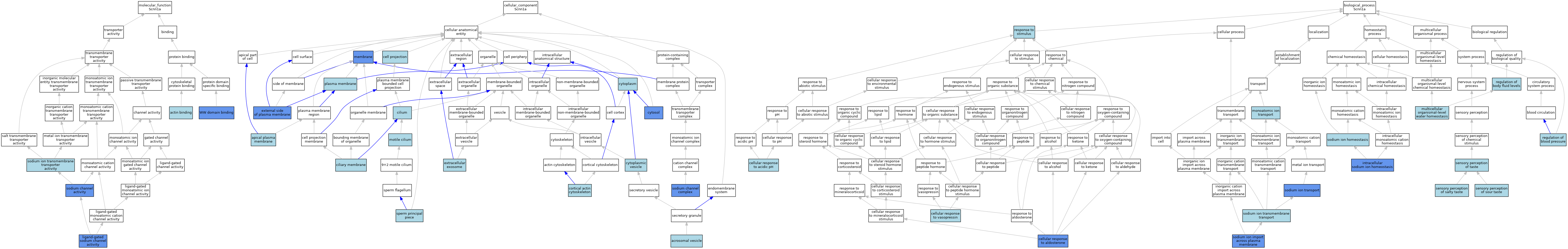
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | IGI | J:126754 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | IGI | J:126754 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | IDA | J:56566 | |||||||||
| Molecular Function | GO:0005272 | sodium channel activity | IDA | J:136460 | |||||||||
| Molecular Function | GO:0015081 | sodium ion transmembrane transporter activity | ISO | J:297461 | |||||||||
| Molecular Function | GO:0050699 | WW domain binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0050699 | WW domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0050699 | WW domain binding | IPI | J:54531 | |||||||||
| Molecular Function | GO:0050699 | WW domain binding | IPI | J:54531 | |||||||||
| Cellular Component | GO:0001669 | acrosomal vesicle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0060170 | ciliary membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005929 | cilium | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030864 | cortical actin cytoskeleton | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:172256 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IDA | J:126754 | |||||||||
| Cellular Component | GO:0070062 | extracellular exosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:172256 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:126754 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:56566 | |||||||||
| Cellular Component | GO:0031514 | motile cilium | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0034706 | sodium channel complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0034706 | sodium channel complex | IDA | J:297464 | |||||||||
| Cellular Component | GO:0034706 | sodium channel complex | IDA | J:286836 | |||||||||
| Cellular Component | GO:0034706 | sodium channel complex | IGI | J:126754 | |||||||||
| Cellular Component | GO:0034706 | sodium channel complex | IGI | J:126754 | |||||||||
| Cellular Component | GO:0097228 | sperm principal piece | ISO | J:155856 | |||||||||
| Biological Process | GO:0071468 | cellular response to acidic pH | ISO | J:164563 | |||||||||
| Biological Process | GO:1904045 | cellular response to aldosterone | NAS | J:320166 | |||||||||
| Biological Process | GO:1904045 | cellular response to aldosterone | IDA | J:160934 | |||||||||
| Biological Process | GO:1904117 | cellular response to vasopressin | NAS | J:320166 | |||||||||
| Biological Process | GO:0006883 | intracellular sodium ion homeostasis | IDA | J:91291 | |||||||||
| Biological Process | GO:0006883 | intracellular sodium ion homeostasis | ISO | J:155856 | |||||||||
| Biological Process | GO:0006883 | intracellular sodium ion homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0050891 | multicellular organismal-level water homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0008217 | regulation of blood pressure | NAS | J:320166 | |||||||||
| Biological Process | GO:0050878 | regulation of body fluid levels | ISO | J:155856 | |||||||||
| Biological Process | GO:0050896 | response to stimulus | IEA | J:60000 | |||||||||
| Biological Process | GO:0050914 | sensory perception of salty taste | NAS | J:320167 | |||||||||
| Biological Process | GO:0050915 | sensory perception of sour taste | NAS | J:320167 | |||||||||
| Biological Process | GO:0050909 | sensory perception of taste | IEA | J:60000 | |||||||||
| Biological Process | GO:0055078 | sodium ion homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0098719 | sodium ion import across plasma membrane | ISO | J:155856 | |||||||||
| Biological Process | GO:0098719 | sodium ion import across plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0098719 | sodium ion import across plasma membrane | IDA | J:91291 | |||||||||
| Biological Process | GO:0035725 | sodium ion transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0035725 | sodium ion transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0006814 | sodium ion transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0006814 | sodium ion transport | IGI | J:126754 | |||||||||
| Biological Process | GO:0006814 | sodium ion transport | IGI | J:136460 | |||||||||
| Biological Process | GO:0006814 | sodium ion transport | IGI | J:136460 | |||||||||
| Biological Process | GO:0006814 | sodium ion transport | IDA | J:56566 | |||||||||
| Biological Process | GO:0006814 | sodium ion transport | IGI | J:126754 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||