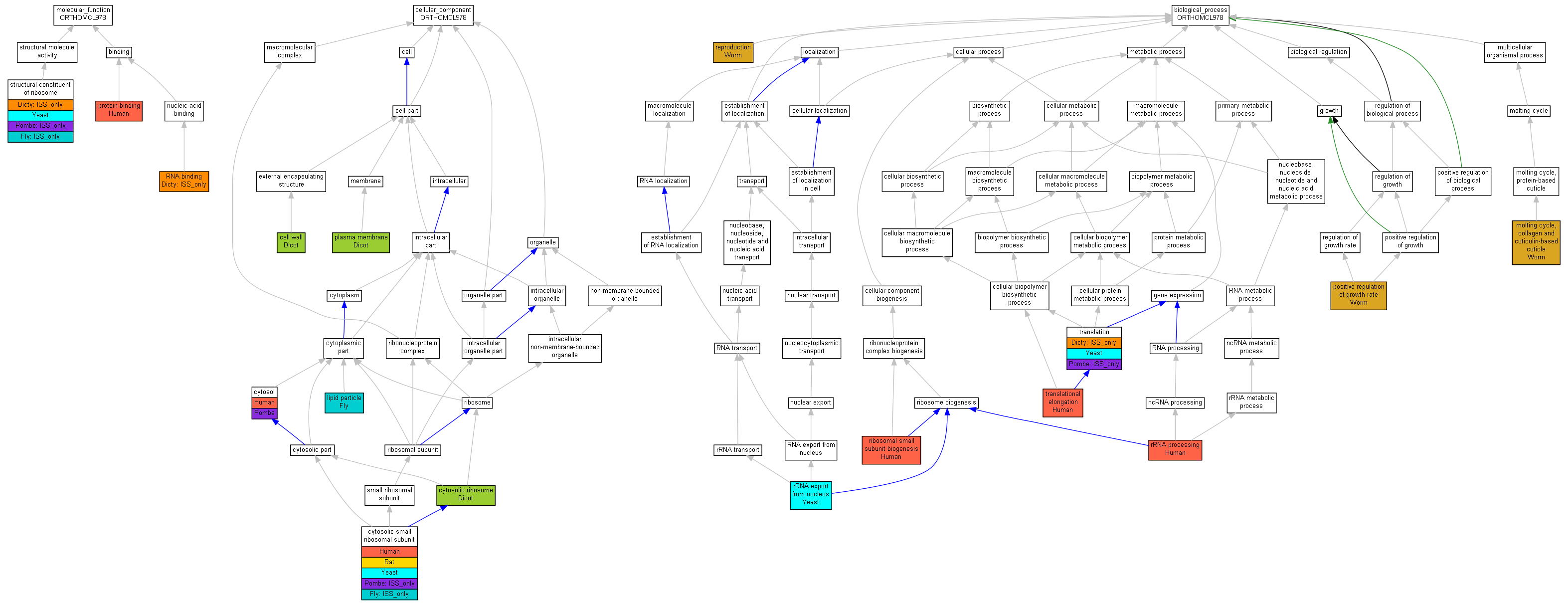
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | X |
| Molecular Function | GO:0005198 | structural molecule activity | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0032991 | macromolecular complex | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | X |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0051234 | establishment of localization | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0051179 | localization | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0000003 | reproduction | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | P62857 | IPI | UniProtKB:Q96BK5 | Human | PMID:17353931 |
| Molecular Function | GO:0003723 | RNA binding | rps28 | ISS | UniProt:P62857 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RpS28a | ISS | NCBI_gi:4506715 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RpS28b | ISS | NCBI_gi:4506715 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | rps28 | ISS | UniProt:P62857 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RPS28A | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RPS28B | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Molecular Function | GO:0003735 | structural constituent of ribosome | rps2801 | ISS | SGD:S000005693 | Pombe | PMID:17072883 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | rps2802 | ISS | SGD:S000005693 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005618 | cell wall | AT3G10090.1 | IDA | Dicot | TAIR:Publication:501718077|PMID:16287169 | |
| Cellular Component | GO:0005618 | cell wall | AT5G03850.1 | IDA | Dicot | TAIR:Publication:501718077|PMID:16287169 | |
| Cellular Component | GO:0005829 | cytosol | P62857 | EXP | Human | PMID:12588972 | |
| Cellular Component | GO:0005829 | cytosol | rps2801 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005829 | cytosol | rps2802 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | RPS28 | IDA | Dicot | TAIR:Publication:501714926|PMID:15734919 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | AT3G10090.1 | IDA | Dicot | TAIR:Publication:501714975|PMID:15821981 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | P62857 | IDA | Human | PMID:8706699 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | P62857 | IDA | Human | PMID:15883184 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | Rps28 | IDA | Rat | RGD:2300010|PMID:947902 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RpS28a | ISS | NCBI_gi:4506715 | Fly | FB:FBrf0105495 |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RpS28b | ISS | NCBI_gi:4506715 | Fly | FB:FBrf0105495 |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RPS28A | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RPS28B | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | rps2801 | ISS | SGD:S000005693 | Pombe | PMID:17072883 |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | rps2802 | ISS | SGD:S000005693 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005811 | lipid particle | RpS28b | IDA | Fly | FB:FBrf0200342 | |
| Cellular Component | GO:0005886 | plasma membrane | AT5G03850.1 | IDA | Dicot | TAIR:Publication:501712339|PMID:15060130 | |
| Biological Process | GO:0018996 | molting cycle, collagen and cuticulin-based cuticle | rps-28 | IMP | WB:WBRNAi00066876|WB:WBPhenotype:0000638 | Worm | WB:WBPaper00026763|PMID:16122351 |
| Biological Process | GO:0040010 | positive regulation of growth rate | rps-28 | IMP | WB:WBRNAi00009323|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0000003 | reproduction | rps-28 | IMP | WB:WBRNAi00009323|WB:WBPhenotype:0000689 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0000003 | reproduction | rps-28 | IMP | WB:WBRNAi00056397|WB:WBPhenotype:0000689 | Worm | WB:WBPaper00025054|PMID:15791247 |
| Biological Process | GO:0042274 | ribosomal small subunit biogenesis | P62857 | IMP | Human | PMID:18697920 | |
| Biological Process | GO:0006407 | rRNA export from nucleus | RPS28A | IGI | SGD:S000004254 | Yeast | SGD_REF:S000087225|PMID:16246728 |
| Biological Process | GO:0006407 | rRNA export from nucleus | RPS28B | IGI | SGD:S000005693 | Yeast | SGD_REF:S000087225|PMID:16246728 |
| Biological Process | GO:0006364 | rRNA processing | P62857 | IMP | Human | PMID:18697920 | |
| Biological Process | GO:0006412 | translation | rps28 | ISS | UniProt:P62857 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006412 | translation | RPS28A | IC | GO:0022627 | Yeast | SGD_REF:S000116584|PMID:3533916 |
| Biological Process | GO:0006412 | translation | RPS28B | IC | GO:0022627 | Yeast | SGD_REF:S000116584|PMID:3533916 |
| Biological Process | GO:0006412 | translation | rps2801 | ISS | SGD:S000005693 | Pombe | PMID:17072883 |
| Biological Process | GO:0006412 | translation | rps2802 | ISS | SGD:S000005693 | Pombe | PMID:17072883 |
| Biological Process | GO:0006414 | translational elongation | P62857 | EXP | Human | PMID:15189156 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |