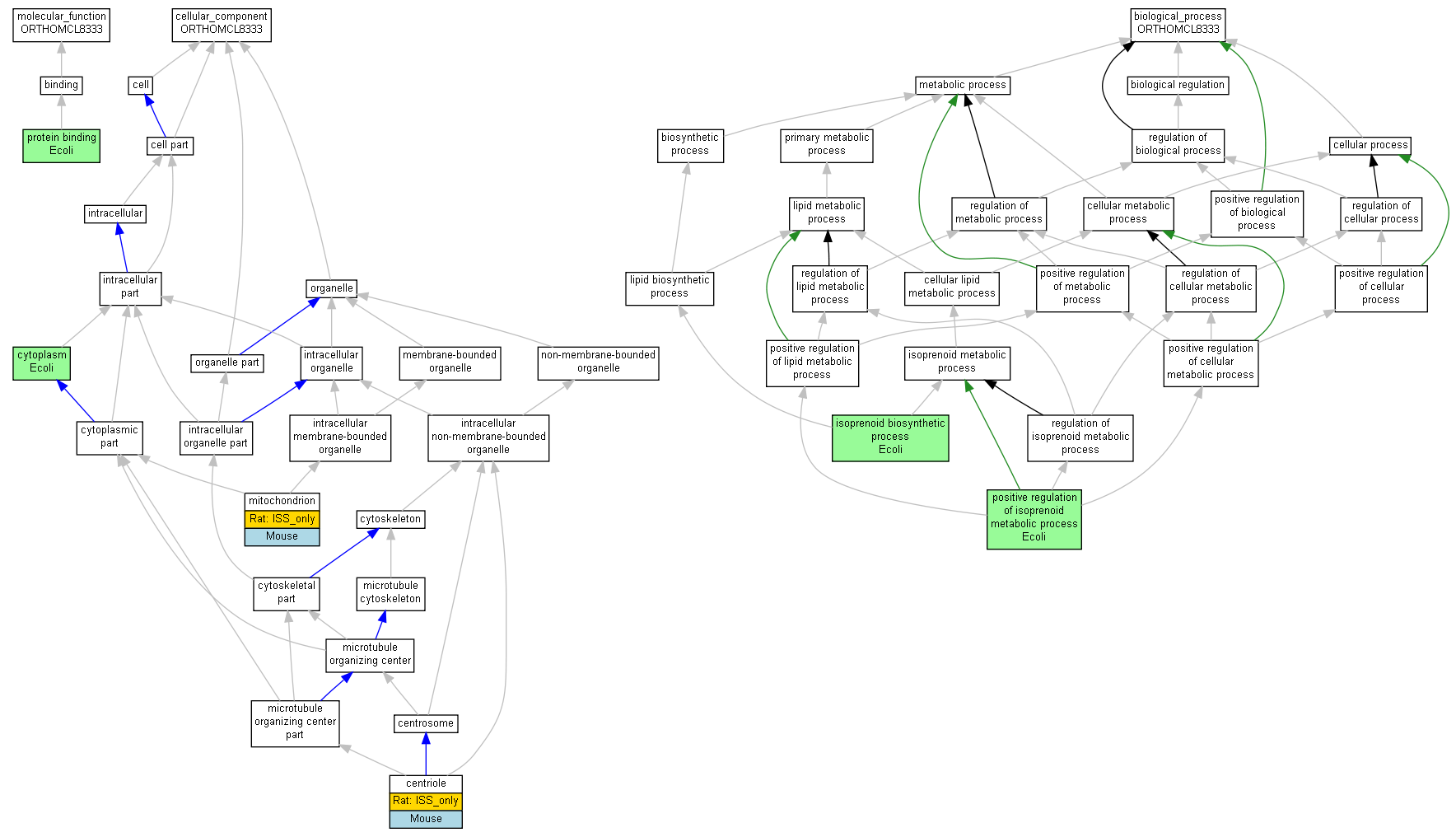
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | X | X | X | X | X | X | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | elbB | IPI | UniProtKB:P0A8V2 | Ecoli | PMID:16606699 |
| Cellular Component | GO:0005814 | centriole | D10Jhu81e | IDA | Mouse | MGI:MGI:3790380|PMID:18239929 | |
| Cellular Component | GO:0005814 | centriole | RGD1303003 | ISO | RGD:1616812 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | elbB | IDA | Ecoli | PMID:16858726 | |
| Cellular Component | GO:0005739 | mitochondrion | D10Jhu81e | IDA | Mouse | MGI:MGI:2682130|PMID:14651853 | |
| Cellular Component | GO:0005739 | mitochondrion | RGD1303003 | ISO | RGD:1616812 | Rat | RGD:1624291 |
| Biological Process | GO:0008299 | isoprenoid biosynthetic process | elbB | IMP | Ecoli | PMID:9603997 | |
| Biological Process | GO:0045828 | positive regulation of isoprenoid metabolic process | elbB | IMP | Ecoli | PMID:9603997 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |