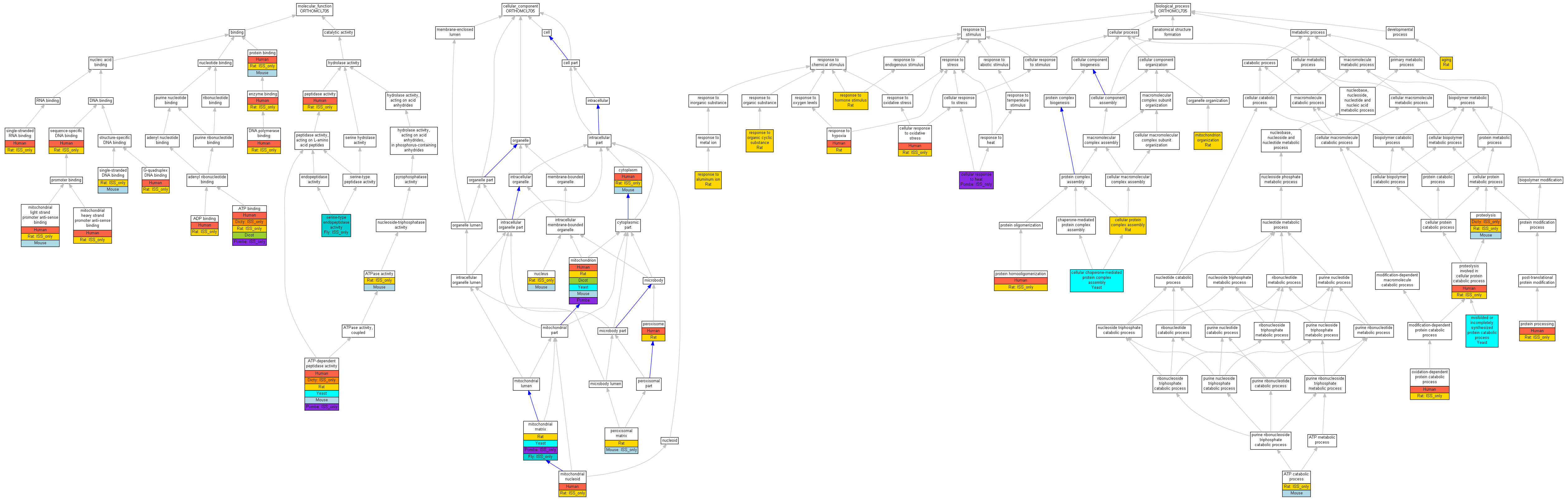
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0031974 | membrane-enclosed lumen | Human | (Mouse) | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044422 | organelle part | Human | (Mouse) | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0010926 | anatomical structure formation | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0043531 | ADP binding | P36776 | IDA | Human | PMID:14739292 | |
| Molecular Function | GO:0043531 | ADP binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0005524 | ATP binding | P36776 | IDA | Human | PMID:14739292 | |
| Molecular Function | GO:0005524 | ATP binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0005524 | ATP binding | DDB_G0278063 | ISS | InterPro:IPR004815 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0005524 | ATP binding | LON1 | IDA | Dicot | TAIR:Publication:501720283|PMID:17137349 | |
| Molecular Function | GO:0005524 | ATP binding | lon1 | ISS | UniProtKB:P36775 | Pombe | PMID:17072883 |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | P36776 | IDA | Human | PMID:8248235 | |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | P36776 | IDA | Human | PMID:17420247 | |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | Lonp1 | IDA | Mouse | MGI:MGI:2655856|PMID:12657466 | |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | Lonp1 | IDA | Rat | RGD:1580665|PMID:12752449 | |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | Lonp1 | ISO | RGD:734454 | Rat | RGD:1624291 |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | DDB_G0278063 | ISS | InterPro:IPR004815 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | PIM1 | IMP | Yeast | SGD_REF:S000040977|PMID:8276800 | |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | PIM1 | IMP | Yeast | SGD_REF:S000061090|PMID:8146662 | |
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | lon1 | ISS | UniProtKB:P36775 | Pombe | PMID:17072883 |
| Molecular Function | GO:0016887 | ATPase activity | Lonp1 | IDA | Mouse | MGI:MGI:2655856|PMID:12657466 | |
| Molecular Function | GO:0016887 | ATPase activity | Lonp1 | ISO | RGD:734454 | Rat | RGD:1624291 |
| Molecular Function | GO:0070182 | DNA polymerase binding | P36776 | IPI | UniProtKB:P54098 | Human | PMID:14739292 |
| Molecular Function | GO:0070182 | DNA polymerase binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0019899 | enzyme binding | Q86WA8 | IPI | UniProtKB:P09110 | Human | PMID:18281296 |
| Molecular Function | GO:0019899 | enzyme binding | Lonp2 | ISO | RGD:1605609 | Rat | RGD:1624291 |
| Molecular Function | GO:0051880 | G-quadruplex DNA binding | P36776 | IDA | Human | PMID:18174225 | |
| Molecular Function | GO:0051880 | G-quadruplex DNA binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0070362 | mitochondrial heavy strand promoter anti-sense binding | P36776 | IDA | Human | PMID:9485316 | |
| Molecular Function | GO:0070362 | mitochondrial heavy strand promoter anti-sense binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0070361 | mitochondrial light strand promoter anti-sense binding | P36776 | IDA | Human | PMID:9485316 | |
| Molecular Function | GO:0070361 | mitochondrial light strand promoter anti-sense binding | Lonp1 | IDA | Mouse | MGI:MGI:2655856|PMID:12657466 | |
| Molecular Function | GO:0070361 | mitochondrial light strand promoter anti-sense binding | Lonp1 | ISO | RGD:734454 | Rat | RGD:1624291 |
| Molecular Function | GO:0070361 | mitochondrial light strand promoter anti-sense binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0008233 | peptidase activity | Q86WA8 | IDA | Human | PMID:18281296 | |
| Molecular Function | GO:0008233 | peptidase activity | Lonp2 | ISO | RGD:1605609 | Rat | RGD:1624291 |
| Molecular Function | GO:0005515 | protein binding | P36776 | IPI | UniProtKB:Q96RR1 | Human | PMID:14739292 |
| Molecular Function | GO:0005515 | protein binding | Lonp2 | IPI | UniProtKB:Q61658 | Mouse | MGI:MGI:3778150|PMID:17931718 |
| Molecular Function | GO:0005515 | protein binding | Lonp2 | ISO | RGD:1332507 | Rat | RGD:1624291 |
| Molecular Function | GO:0005515 | protein binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | P36776 | IDA | Human | PMID:14739292 | |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | CG8798 | ISS | UniProtKB:P93655 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | CG8798 | ISS | UniProtKB:P36776 | Fly | FB:FBrf0159903 |
| Molecular Function | GO:0003697 | single-stranded DNA binding | Lonp1 | IDA | Mouse | MGI:MGI:2655856|PMID:12657466 | |
| Molecular Function | GO:0003697 | single-stranded DNA binding | Lonp1 | ISO | RGD:734454 | Rat | RGD:1624291 |
| Molecular Function | GO:0003727 | single-stranded RNA binding | P36776 | IDA | Human | PMID:14739292 | |
| Molecular Function | GO:0003727 | single-stranded RNA binding | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | P36776 | IDA | Human | PMID:18029348 | |
| Cellular Component | GO:0005737 | cytoplasm | Lonp2 | IDA | Mouse | MGI:MGI:3778150|PMID:17931718 | |
| Cellular Component | GO:0005737 | cytoplasm | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | Lonp2 | ISO | RGD:1332507 | Rat | RGD:1624291 |
| Cellular Component | GO:0005759 | mitochondrial matrix | Lonp1 | IDA | Rat | RGD:2303495|PMID:15560797 | |
| Cellular Component | GO:0005759 | mitochondrial matrix | CG8798 | ISS | UniProtKB:P36776 | Fly | FB:FBrf0159903 |
| Cellular Component | GO:0005759 | mitochondrial matrix | PIM1 | IMP | Yeast | SGD_REF:S000040977|PMID:8276800 | |
| Cellular Component | GO:0005759 | mitochondrial matrix | PIM1 | IMP | Yeast | SGD_REF:S000061090|PMID:8146662 | |
| Cellular Component | GO:0005759 | mitochondrial matrix | lon1 | ISS | UniProtKB:P36775 | Pombe | PMID:17072883 |
| Cellular Component | GO:0042645 | mitochondrial nucleoid | P36776 | IDA | Human | PMID:18063578 | |
| Cellular Component | GO:0042645 | mitochondrial nucleoid | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | P36776 | IDA | Human | PMID:8248235 | |
| Cellular Component | GO:0005739 | mitochondrion | Lonp1 | IDA | Mouse | MGI:MGI:2682130|PMID:14651853 | |
| Cellular Component | GO:0005739 | mitochondrion | Lonp1 | IDA | Mouse | MGI:MGI:2655856|PMID:12657466 | |
| Cellular Component | GO:0005739 | mitochondrion | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | Lonp1 | IDA | Rat | RGD:1580665|PMID:12752449 | |
| Cellular Component | GO:0005739 | mitochondrion | Lonp1 | ISO | RGD:734454 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | LON1 | IDA | Dicot | TAIR:Publication:501720283|PMID:17137349 | |
| Cellular Component | GO:0005739 | mitochondrion | PIM1 | IDA | Yeast | SGD_REF:S000117178|PMID:16823961 | |
| Cellular Component | GO:0005739 | mitochondrion | lon1 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005634 | nucleus | Lonp2 | IDA | Mouse | MGI:MGI:3778150|PMID:17931718 | |
| Cellular Component | GO:0005634 | nucleus | Lonp2 | ISO | RGD:1332507 | Rat | RGD:1624291 |
| Cellular Component | GO:0005782 | peroxisomal matrix | Lonp2 | ISS | UniProtKB:Q6TXI3 | Mouse | MGI:MGI:3693380|PMID:14561759 |
| Cellular Component | GO:0005782 | peroxisomal matrix | Lonp2 | IDA | Rat | RGD:14561759 | |
| Cellular Component | GO:0005782 | peroxisomal matrix | Lonp2 | IDA | Rat | RGD:1580664|PMID:14561759 | |
| Cellular Component | GO:0005777 | peroxisome | Q86WA8 | IDA | Human | PMID:18281296 | |
| Cellular Component | GO:0005777 | peroxisome | Lonp2 | ISO | RGD:1605609 | Rat | RGD:1624291 |
| Cellular Component | GO:0005777 | peroxisome | Lonp1 | IDA | Rat | RGD:1580664|PMID:14561759 | |
| Biological Process | GO:0007568 | aging | Lonp1 | IEP | Rat | RGD:2303495|PMID:15560797 | |
| Biological Process | GO:0006200 | ATP catabolic process | Lonp1 | IDA | Mouse | MGI:MGI:2655856|PMID:12657466 | |
| Biological Process | GO:0006200 | ATP catabolic process | Lonp1 | ISO | RGD:734454 | Rat | RGD:1624291 |
| Biological Process | GO:0034619 | cellular chaperone-mediated protein complex assembly | PIM1 | IGI | SGD:S000004695|SGD:S000000819 | Yeast | SGD_REF:S000040612|PMID:8810243 |
| Biological Process | GO:0043623 | cellular protein complex assembly | Lonp1 | IMP | Rat | RGD:633879|PMID:12082077 | |
| Biological Process | GO:0034605 | cellular response to heat | lon1 | ISS | UniProtKB:P36775 | Pombe | PMID:17072883 |
| Biological Process | GO:0034599 | cellular response to oxidative stress | P36776 | IC | GO:0019941 | Human | PMID:12198491 |
| Biological Process | GO:0034599 | cellular response to oxidative stress | P36776 | IDA | Human | PMID:17420247 | |
| Biological Process | GO:0034599 | cellular response to oxidative stress | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Biological Process | GO:0006515 | misfolded or incompletely synthesized protein catabolic process | PIM1 | IMP | Yeast | SGD_REF:S000044484|PMID:7957078 | |
| Biological Process | GO:0006515 | misfolded or incompletely synthesized protein catabolic process | PIM1 | IGI | SGD:S000003806 | Yeast | SGD_REF:S000044484|PMID:7957078 |
| Biological Process | GO:0006515 | misfolded or incompletely synthesized protein catabolic process | PIM1 | IMP | Yeast | SGD_REF:S000129400|PMID:16336126 | |
| Biological Process | GO:0007005 | mitochondrion organization | Lonp1 | IEP | Rat | RGD:1580666|PMID:10050756 | |
| Biological Process | GO:0070407 | oxidation-dependent protein catabolic process | P36776 | IMP | Human | PMID:12198491 | |
| Biological Process | GO:0070407 | oxidation-dependent protein catabolic process | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Biological Process | GO:0051260 | protein homooligomerization | P36776 | IDA | Human | PMID:14739292 | |
| Biological Process | GO:0051260 | protein homooligomerization | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Biological Process | GO:0016485 | protein processing | Q86WA8 | IMP | Human | PMID:18281296 | |
| Biological Process | GO:0016485 | protein processing | Lonp2 | ISO | RGD:1605609 | Rat | RGD:1624291 |
| Biological Process | GO:0006508 | proteolysis | Lonp1 | IDA | Mouse | MGI:MGI:2655856|PMID:12657466 | |
| Biological Process | GO:0006508 | proteolysis | Lonp1 | ISO | RGD:734454 | Rat | RGD:1624291 |
| Biological Process | GO:0006508 | proteolysis | DDB_G0278063 | ISS | InterPro:IPR004815 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0051603 | proteolysis involved in cellular protein catabolic process | P36776 | IDA | Human | PMID:8248235 | |
| Biological Process | GO:0051603 | proteolysis involved in cellular protein catabolic process | P36776 | IDA | Human | PMID:17420247 | |
| Biological Process | GO:0051603 | proteolysis involved in cellular protein catabolic process | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Biological Process | GO:0010044 | response to aluminum ion | Lonp1 | IEP | Rat | RGD:2303407|PMID:19010380 | |
| Biological Process | GO:0009725 | response to hormone stimulus | Lonp1 | IEP | Rat | RGD:1580666|PMID:10050756 | |
| Biological Process | GO:0001666 | response to hypoxia | P36776 | IEP | Human | PMID:17418790 | |
| Biological Process | GO:0001666 | response to hypoxia | Lonp1 | IEP | Rat | RGD:633879|PMID:12082077 | |
| Biological Process | GO:0001666 | response to hypoxia | Lonp1 | ISO | RGD:734453 | Rat | RGD:1624291 |
| Biological Process | GO:0014070 | response to organic cyclic substance | Lonp2 | IEP | Rat | RGD:2303408|PMID:17929048 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |