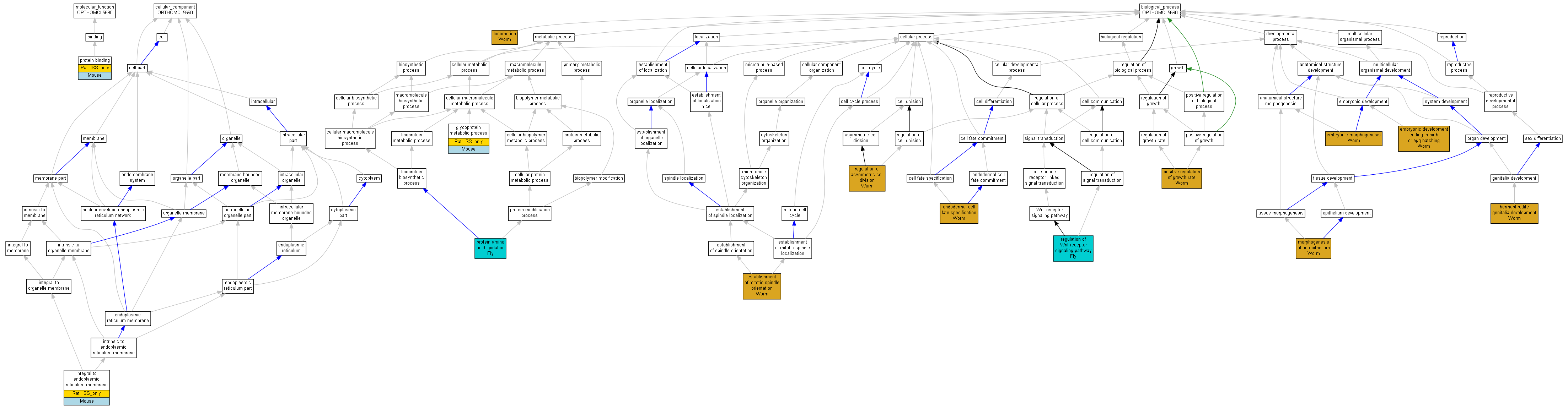
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0051234 | establishment of localization | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0051179 | localization | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0040011 | locomotion | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0000003 | reproduction | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Biological Process | GO:0022414 | reproductive process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | X | X | X | X | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | Porcn | IPI | UniProtKB:P04426|UniProtKB:P17553|UniProtKB:P27467|UniProtKB:P22724|UniProtKB:P22725|UniProtKB:P22726|UniProtKB:P22727|UniProtKB:P24383|UniProtKB:P28047 | Mouse | MGI:MGI:1860653|PMID:10866835 |
| Molecular Function | GO:0005515 | protein binding | Porcn | ISO | RGD:1557351 | Rat | RGD:1624291 |
| Cellular Component | GO:0030176 | integral to endoplasmic reticulum membrane | Porcn | IDA | Mouse | MGI:MGI:1860653|PMID:10866835 | |
| Cellular Component | GO:0030176 | integral to endoplasmic reticulum membrane | Porcn | ISO | RGD:1557351 | Rat | RGD:1624291 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | mom-1 | IMP | WB:se2|WB:ne117|WB:zu188|WB:zu204|WB:zu237 | Worm | WB:WBPaper00002871 |
| Biological Process | GO:0048598 | embryonic morphogenesis | mom-1 | IMP | WB:se2|WB:ne117|WB:zu188|WB:zu204|WB:zu237 | Worm | WB:WBPaper00002871 |
| Biological Process | GO:0001714 | endodermal cell fate specification | mom-1 | IMP | WB:or10|WB:or46|WB:or65|WB:or83|WB:or70 | Worm | WB:WBPaper00002870 |
| Biological Process | GO:0001714 | endodermal cell fate specification | mom-1 | IMP | WB:se2|WB:ne117|WB:zu188|WB:zu204|WB:zu237 | Worm | WB:WBPaper00002871 |
| Biological Process | GO:0000132 | establishment of mitotic spindle orientation | mom-1 | IMP | WB:or10|WB:or46|WB:or70 | Worm | WB:WBPaper00002870 |
| Biological Process | GO:0000132 | establishment of mitotic spindle orientation | mom-1 | IMP | Worm | WB:WBPaper00002871 | |
| Biological Process | GO:0009100 | glycoprotein metabolic process | Porcn | IDA | Mouse | MGI:MGI:1860653|PMID:10866835 | |
| Biological Process | GO:0009100 | glycoprotein metabolic process | Porcn | ISO | RGD:1557351 | Rat | RGD:1624291 |
| Biological Process | GO:0040035 | hermaphrodite genitalia development | mom-1 | IMP | WB:or10|WB:WBPhenotype:0000697 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040035 | hermaphrodite genitalia development | mom-1 | IMP | WB:or46|WB:WBPhenotype:0000697 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040035 | hermaphrodite genitalia development | mom-1 | IMP | WB:or65|WB:WBPhenotype:0000697 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040035 | hermaphrodite genitalia development | mom-1 | IMP | WB:or70|WB:WBPhenotype:0000697 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040035 | hermaphrodite genitalia development | mom-1 | IMP | WB:or83|WB:WBPhenotype:0000697 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040011 | locomotion | mom-1 | IMP | WB:or10|WB:WBPhenotype:0000643 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040011 | locomotion | mom-1 | IMP | WB:or46|WB:WBPhenotype:0000643 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040011 | locomotion | mom-1 | IMP | WB:or65|WB:WBPhenotype:0000643 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040011 | locomotion | mom-1 | IMP | WB:or70|WB:WBPhenotype:0000643 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040011 | locomotion | mom-1 | IMP | WB:or83|WB:WBPhenotype:0000643 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0002009 | morphogenesis of an epithelium | mom-1 | IMP | WB:or10|WB:WBPhenotype:0000038 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0002009 | morphogenesis of an epithelium | mom-1 | IMP | WB:or46|WB:WBPhenotype:0000038 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0002009 | morphogenesis of an epithelium | mom-1 | IMP | WB:or65|WB:WBPhenotype:0000038 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0002009 | morphogenesis of an epithelium | mom-1 | IMP | WB:or70|WB:WBPhenotype:0000038 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0002009 | morphogenesis of an epithelium | mom-1 | IMP | WB:or83|WB:WBPhenotype:0000038 | Worm | WB:WBPaper00002870|PMID:9288749 |
| Biological Process | GO:0040010 | positive regulation of growth rate | mom-1 | IMP | WB:WBRNAi00067937|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00031036|PMID:17897480 |
| Biological Process | GO:0040010 | positive regulation of growth rate | mom-1 | IMP | WB:WBRNAi00068239|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00031036|PMID:17897480 |
| Biological Process | GO:0006497 | protein amino acid lipidation | por | IMP | Fly | FB:FBrf0179536|PMID:15166250 | |
| Biological Process | GO:0009786 | regulation of asymmetric cell division | mom-1 | IMP | WB:se2 | Worm | WB:WBPaper00002871 |
| Biological Process | GO:0030111 | regulation of Wnt receptor signaling pathway | por | IMP | Fly | FB:FBrf0147096|PMID:11821428 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |