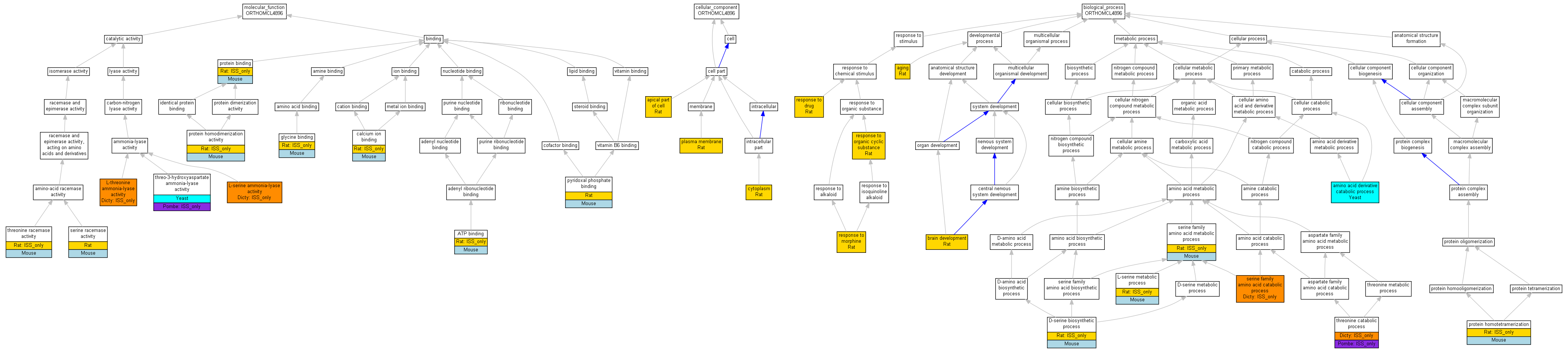
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | X | X | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | X | X | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | X | X | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | X | X | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0010926 | anatomical structure formation | Human | Mouse | (Rat) | Chicken | X | X | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | X | X | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | X | X | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | X | X | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | X | X | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | X | X | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | Srr | IDA | Mouse | MGI:MGI:3760217|PMID:17293453 | |
| Molecular Function | GO:0005524 | ATP binding | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Molecular Function | GO:0005509 | calcium ion binding | Srr | IDA | Mouse | MGI:MGI:2183617|PMID:12021263 | |
| Molecular Function | GO:0005509 | calcium ion binding | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Molecular Function | GO:0016594 | glycine binding | Srr | IDA | Mouse | MGI:MGI:3529732|PMID:15710237 | |
| Molecular Function | GO:0016594 | glycine binding | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Molecular Function | GO:0003941 | L-serine ammonia-lyase activity | DDB_G0289463 | ISS | SGD:S000000569 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004794 | L-threonine ammonia-lyase activity | DDB_G0289463 | ISS | SGD:S000000569 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0005515 | protein binding | Srr | IPI | UniProtKB:Q9QZX7 | Mouse | MGI:MGI:2183617|PMID:12021263 |
| Molecular Function | GO:0005515 | protein binding | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Molecular Function | GO:0042803 | protein homodimerization activity | Srr | IDA | Mouse | MGI:MGI:2183617|PMID:12021263 | |
| Molecular Function | GO:0042803 | protein homodimerization activity | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Molecular Function | GO:0030170 | pyridoxal phosphate binding | Srr | IMP | Mouse | MGI:MGI:1347751|PMID:10557334 | |
| Molecular Function | GO:0030170 | pyridoxal phosphate binding | Srr | IDA | Rat | RGD:2302218|PMID:9892700 | |
| Molecular Function | GO:0030170 | pyridoxal phosphate binding | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Molecular Function | GO:0030378 | serine racemase activity | Srr | IDA | Mouse | MGI:MGI:3760217|PMID:17293453 | |
| Molecular Function | GO:0030378 | serine racemase activity | Srr | IDA | Mouse | MGI:MGI:3529611|PMID:15536068 | |
| Molecular Function | GO:0030378 | serine racemase activity | Srr | IDA | Mouse | MGI:MGI:1347751|PMID:10557334 | |
| Molecular Function | GO:0030378 | serine racemase activity | Srr | IDA | Mouse | MGI:MGI:3529732|PMID:15710237 | |
| Molecular Function | GO:0030378 | serine racemase activity | Srr | ISO | NCBI:NP_068766 | Mouse | MGI:MGI:1926659|PMID:11054547 |
| Molecular Function | GO:0030378 | serine racemase activity | Srr | IDA | Mouse | MGI:MGI:2183617|PMID:12021263 | |
| Molecular Function | GO:0030378 | serine racemase activity | Srr | IDA | Rat | RGD:2302218|PMID:9892700 | |
| Molecular Function | GO:0030378 | serine racemase activity | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Molecular Function | GO:0030848 | threo-3-hydroxyaspartate ammonia-lyase activity | SRY1 | IDA | Yeast | SGD_REF:S000076238|PMID:12951240 | |
| Molecular Function | GO:0030848 | threo-3-hydroxyaspartate ammonia-lyase activity | SRY1 | IMP | Yeast | SGD_REF:S000076238|PMID:12951240 | |
| Molecular Function | GO:0030848 | threo-3-hydroxyaspartate ammonia-lyase activity | SPCC320.14 | ISS | UniProtKB:P36007 | Pombe | PMID:17072883 |
| Molecular Function | GO:0018114 | threonine racemase activity | Srr | IDA | Mouse | MGI:MGI:3529611|PMID:15536068 | |
| Molecular Function | GO:0018114 | threonine racemase activity | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Cellular Component | GO:0045177 | apical part of cell | Srr | IDA | Rat | RGD:2302217|PMID:16289842 | |
| Cellular Component | GO:0005737 | cytoplasm | Srr | IDA | Rat | RGD:2302217|PMID:16289842 | |
| Cellular Component | GO:0005886 | plasma membrane | Srr | IDA | Rat | RGD:2302217|PMID:16289842 | |
| Biological Process | GO:0007568 | aging | Srr | IEP | Rat | RGD:2302214|PMID:16842499 | |
| Biological Process | GO:0042219 | amino acid derivative catabolic process | SRY1 | IDA | Yeast | SGD_REF:S000076238|PMID:12951240 | |
| Biological Process | GO:0042219 | amino acid derivative catabolic process | SRY1 | IMP | Yeast | SGD_REF:S000076238|PMID:12951240 | |
| Biological Process | GO:0007420 | brain development | Srr | IEP | Rat | RGD:2302215|PMID:16739185 | |
| Biological Process | GO:0070179 | D-serine biosynthetic process | Srr | IDA | Mouse | MGI:MGI:2183617|PMID:12021263 | |
| Biological Process | GO:0070179 | D-serine biosynthetic process | Srr | IDA | Mouse | MGI:MGI:1926659|PMID:11054547 | |
| Biological Process | GO:0070179 | D-serine biosynthetic process | Srr | IDA | Mouse | MGI:MGI:3529732|PMID:15710237 | |
| Biological Process | GO:0070179 | D-serine biosynthetic process | Srr | IDA | Mouse | MGI:MGI:1347751|PMID:10557334 | |
| Biological Process | GO:0070179 | D-serine biosynthetic process | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Biological Process | GO:0006563 | L-serine metabolic process | Srr | IDA | Mouse | MGI:MGI:3769629|PMID:16517698 | |
| Biological Process | GO:0006563 | L-serine metabolic process | Srr | IDA | Mouse | MGI:MGI:1347751|PMID:10557334 | |
| Biological Process | GO:0006563 | L-serine metabolic process | Srr | IDA | Mouse | MGI:MGI:3529732|PMID:15710237 | |
| Biological Process | GO:0006563 | L-serine metabolic process | Srr | IDA | Mouse | MGI:MGI:2183617|PMID:12021263 | |
| Biological Process | GO:0006563 | L-serine metabolic process | Srr | ISO | NCBI:NP_068766 | Mouse | MGI:MGI:1926659|PMID:11054547 |
| Biological Process | GO:0006563 | L-serine metabolic process | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Biological Process | GO:0051289 | protein homotetramerization | Srr | IDA | Mouse | MGI:MGI:2183617|PMID:12021263 | |
| Biological Process | GO:0051289 | protein homotetramerization | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Biological Process | GO:0042493 | response to drug | Srr | IEP | Rat | RGD:2302213|PMID:17109841 | |
| Biological Process | GO:0043278 | response to morphine | Srr | IEP | Rat | RGD:2302212|PMID:18603832 | |
| Biological Process | GO:0014070 | response to organic cyclic substance | Srr | IEP | Rat | RGD:2302216|PMID:16716293 | |
| Biological Process | GO:0009071 | serine family amino acid catabolic process | DDB_G0289463 | ISS | SGD:S000000569 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0009069 | serine family amino acid metabolic process | Srr | IDA | Mouse | MGI:MGI:3529611|PMID:15536068 | |
| Biological Process | GO:0009069 | serine family amino acid metabolic process | Srr | ISO | RGD:1550738 | Rat | RGD:1624291 |
| Biological Process | GO:0006567 | threonine catabolic process | DDB_G0289463 | ISS | SGD:S000000569 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006567 | threonine catabolic process | SPCC320.14 | ISS | UniProtKB:P36007 | Pombe | PMID:17072883 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |