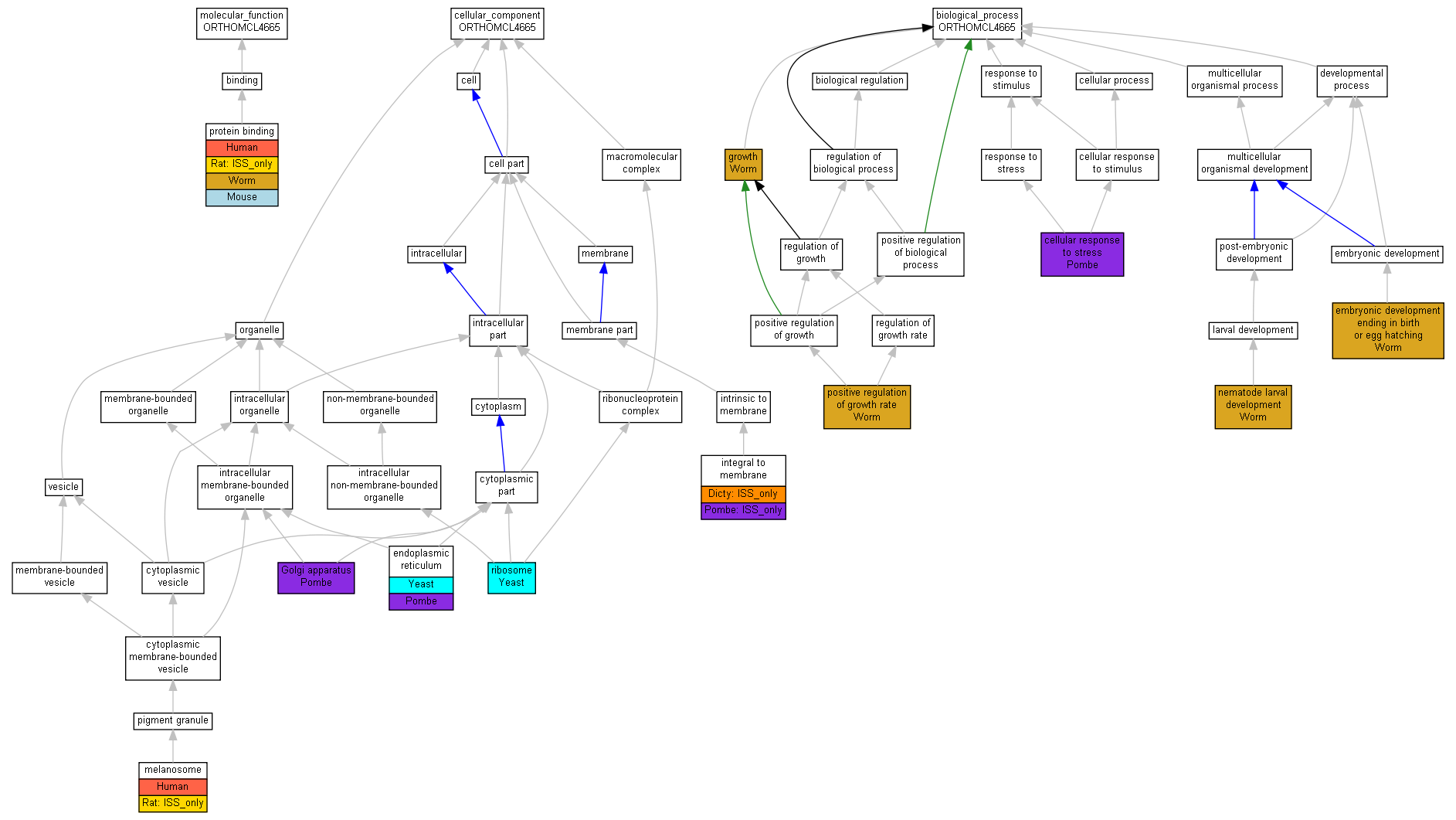
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | (Dicty) | X | Yeast | Pombe | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | (Dicty) | X | Yeast | Pombe | X |
| Cellular Component | GO:0032991 | macromolecular complex | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | X | Yeast | Pombe | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | P57088 | IPI | UniProtKB:O75365 | Human | PMID:17353931 |
| Molecular Function | GO:0005515 | protein binding | P57088 | IPI | UniProtKB:P62140 | Human | PMID:17353931 |
| Molecular Function | GO:0005515 | protein binding | P57088 | IPI | UniProtKB:Q9HAW0 | Human | PMID:17353931 |
| Molecular Function | GO:0005515 | protein binding | P57088 | IPI | UniProtKB:Q99608 | Human | PMID:17353931 |
| Molecular Function | GO:0005515 | protein binding | Tmem33 | IPI | UniProtKB:Q3V4A2 | Mouse | MGI:MGI:3609918|PMID:11591653 |
| Molecular Function | GO:0005515 | protein binding | Tmem33 | ISO | RGD:733060 | Rat | RGD:1624291 |
| Molecular Function | GO:0005515 | protein binding | Tmem33 | ISO | RGD:1353146 | Rat | RGD:1624291 |
| Molecular Function | GO:0005515 | protein binding | Y37D8A.17 | IPI | UniProtKB:Q18529 | Worm | PMID:14704431 |
| Cellular Component | GO:0005783 | endoplasmic reticulum | YLL023C | IDA | Yeast | SGD_REF:S000074185|PMID:14562095 | |
| Cellular Component | GO:0005783 | endoplasmic reticulum | YLR064W | IDA | Yeast | SGD_REF:S000074185|PMID:14562095 | |
| Cellular Component | GO:0005783 | endoplasmic reticulum | SPBC1539.04 | ISS | SGD:S000003946 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005783 | endoplasmic reticulum | SPBC1539.04 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005794 | Golgi apparatus | SPBC1539.04 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0016021 | integral to membrane | tmem33 | ISS | CBS:TMHMM | Dicty | dictyBase_REF:10155 |
| Cellular Component | GO:0016021 | integral to membrane | SPBC1539.04 | ISS | CBS:TMHMM | Pombe | GOC:unpublished |
| Cellular Component | GO:0042470 | melanosome | P57088 | IDA | Human | PMID:17081065 | |
| Cellular Component | GO:0042470 | melanosome | Tmem33 | ISO | RGD:1353146 | Rat | RGD:1624291 |
| Cellular Component | GO:0005840 | ribosome | YLL023C | IDA | Yeast | SGD_REF:S000115662|PMID:16702403 | |
| Biological Process | GO:0033554 | cellular response to stress | SPBC1539.04 | IEP | GOC:pombekw2GO | Pombe | PMID:12529438 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | Y37D8A.17 | IMP | WB:WBRNAi00009304|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0040007 | growth | Y37D8A.17 | IMP | WB:WBRNAi00026649|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0002119 | nematode larval development | Y37D8A.17 | IMP | WB:WBRNAi00026649|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0040010 | positive regulation of growth rate | Y37D8A.17 | IMP | WB:WBRNAi00009304|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00005654|PMID:12529635 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |