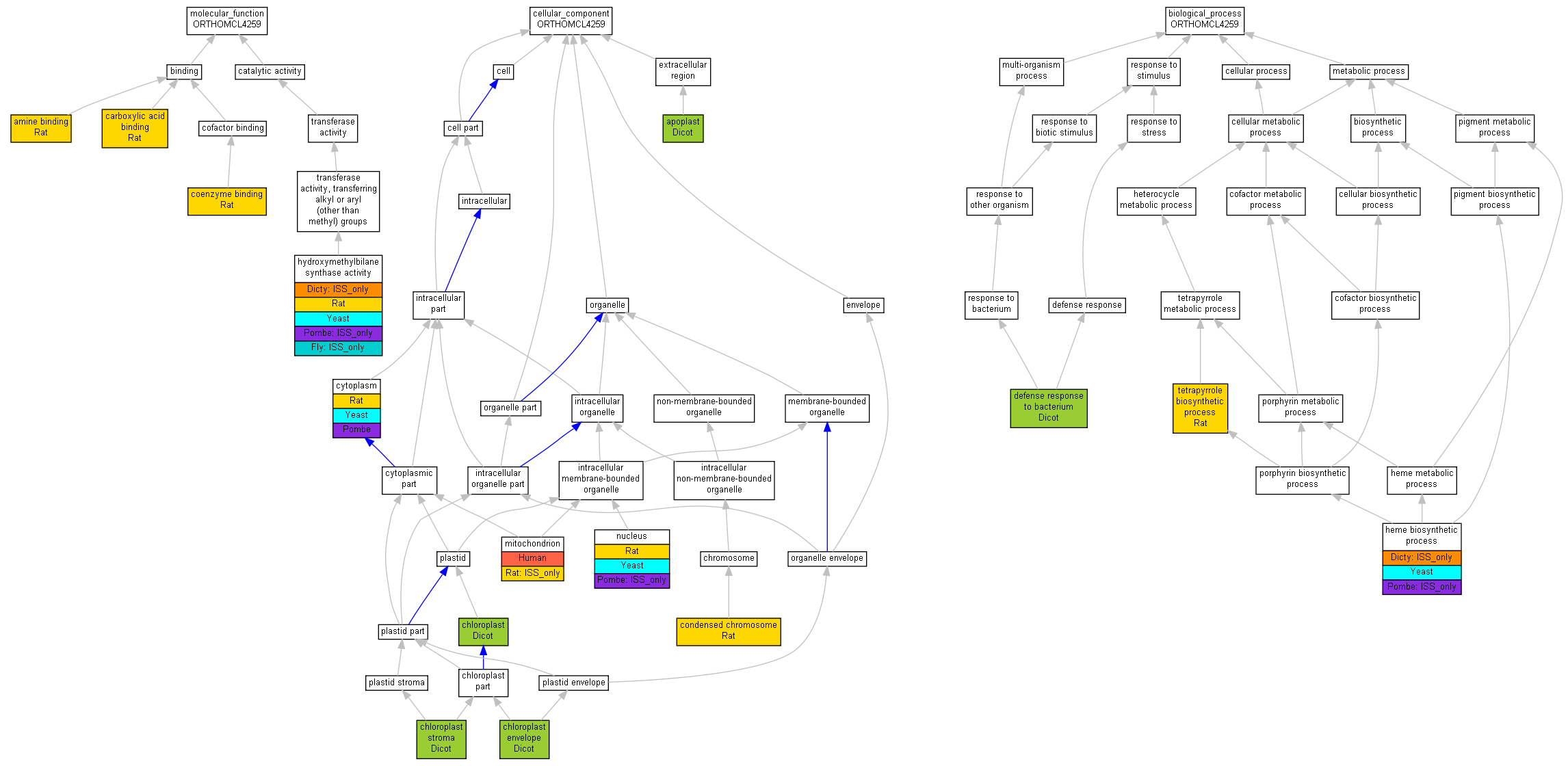
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | Zfish | (Fly) | X | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0031975 | envelope | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0005576 | extracellular region | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | Fly | X | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | Fly | X | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0051704 | multi-organism process | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0043176 | amine binding | Hmbs | IPI | ChEBI:17381 | Rat | RGD:2301695|PMID:9344466 |
| Molecular Function | GO:0031406 | carboxylic acid binding | Hmbs | IPI | ChEBI:17381 | Rat | RGD:2301695|PMID:9344466 |
| Molecular Function | GO:0050662 | coenzyme binding | Hmbs | IPI | ChEBI:42121 | Rat | RGD:2301858|PMID:18760440 |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | Hmbs | IDA | Rat | RGD:2301695|PMID:9344466 | |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | Hmbs | IDA | Rat | RGD:2301704|PMID:6712591 | |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | Hmbs | IDA | Rat | RGD:2301691|PMID:12127573 | |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | Hmbs | IMP | Rat | RGD:2301858|PMID:18760440 | |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | l(3)02640 | ISS | UniProtKB:P19356 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | hemC | ISS | SGD:S000002364 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | HEM3 | IMP | Yeast | SGD_REF:S000046021|PMID:323256 | |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | HEM3 | IEP | Yeast | SGD_REF:S000045327|PMID:1508149 | |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | HEM3 | IMP | Yeast | SGD_REF:S000045327|PMID:1508149 | |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | HEM3 | ISS | Yeast | SGD_REF:S000045327|PMID:1508149 | |
| Molecular Function | GO:0004418 | hydroxymethylbilane synthase activity | hem3 | ISS | UniProtKB:P28789 | Pombe | PMID:17072883 |
| Cellular Component | GO:0048046 | apoplast | HEMC | IDA | Dicot | TAIR:Publication:501725189|PMID:18538804 | |
| Cellular Component | GO:0009507 | chloroplast | HEMC | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0009941 | chloroplast envelope | HEMC | IDA | Dicot | TAIR:Publication:501710686|PMID:12938931 | |
| Cellular Component | GO:0009570 | chloroplast stroma | HEMC | IDA | Dicot | TAIR:Publication:501728638|PMID:16207701 | |
| Cellular Component | GO:0009570 | chloroplast stroma | HEMC | IDA | Dicot | TAIR:Publication:501727324|PMID:18633119 | |
| Cellular Component | GO:0000793 | condensed chromosome | Hmbs | IDA | Rat | RGD:2301689|PMID:12917623 | |
| Cellular Component | GO:0005737 | cytoplasm | Hmbs | IDA | Rat | RGD:2301689|PMID:12917623 | |
| Cellular Component | GO:0005737 | cytoplasm | HEM3 | IDA | Yeast | SGD_REF:S000074185|PMID:14562095 | |
| Cellular Component | GO:0005737 | cytoplasm | hem3 | ISS | UniProtKB:P28789 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005737 | cytoplasm | hem3 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005739 | mitochondrion | P08397 | IDA | Human | PMID:18029348 | |
| Cellular Component | GO:0005739 | mitochondrion | Hmbs | ISO | RGD:733095 | Rat | RGD:1624291 |
| Cellular Component | GO:0005634 | nucleus | Hmbs | IDA | Rat | RGD:2301692|PMID:11953837 | |
| Cellular Component | GO:0005634 | nucleus | Hmbs | IDA | Rat | RGD:2301689|PMID:12917623 | |
| Cellular Component | GO:0005634 | nucleus | HEM3 | IDA | Yeast | SGD_REF:S000074185|PMID:14562095 | |
| Cellular Component | GO:0005634 | nucleus | hem3 | ISS | UniProtKB:P28789 | Pombe | PMID:17072883 |
| Biological Process | GO:0042742 | defense response to bacterium | HEMC | IEP | Dicot | TAIR:Publication:501720163|PMID:17028151 | |
| Biological Process | GO:0006783 | heme biosynthetic process | hemC | ISS | SGD:S000002364 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006783 | heme biosynthetic process | HEM3 | IMP | Yeast | SGD_REF:S000046021|PMID:323256 | |
| Biological Process | GO:0006783 | heme biosynthetic process | hem3 | ISS | UniProtKB:P28789 | Pombe | PMID:17072883 |
| Biological Process | GO:0033014 | tetrapyrrole biosynthetic process | Hmbs | IMP | Rat | RGD:2301858|PMID:18760440 | |
| Biological Process | GO:0033014 | tetrapyrrole biosynthetic process | Hmbs | IDA | Rat | RGD:2301704|PMID:6712591 | |
| Biological Process | GO:0033014 | tetrapyrrole biosynthetic process | Hmbs | IDA | Rat | RGD:2301695|PMID:9344466 | |
| Biological Process | GO:0033014 | tetrapyrrole biosynthetic process | Hmbs | IDA | Rat | RGD:2301691|PMID:12127573 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |