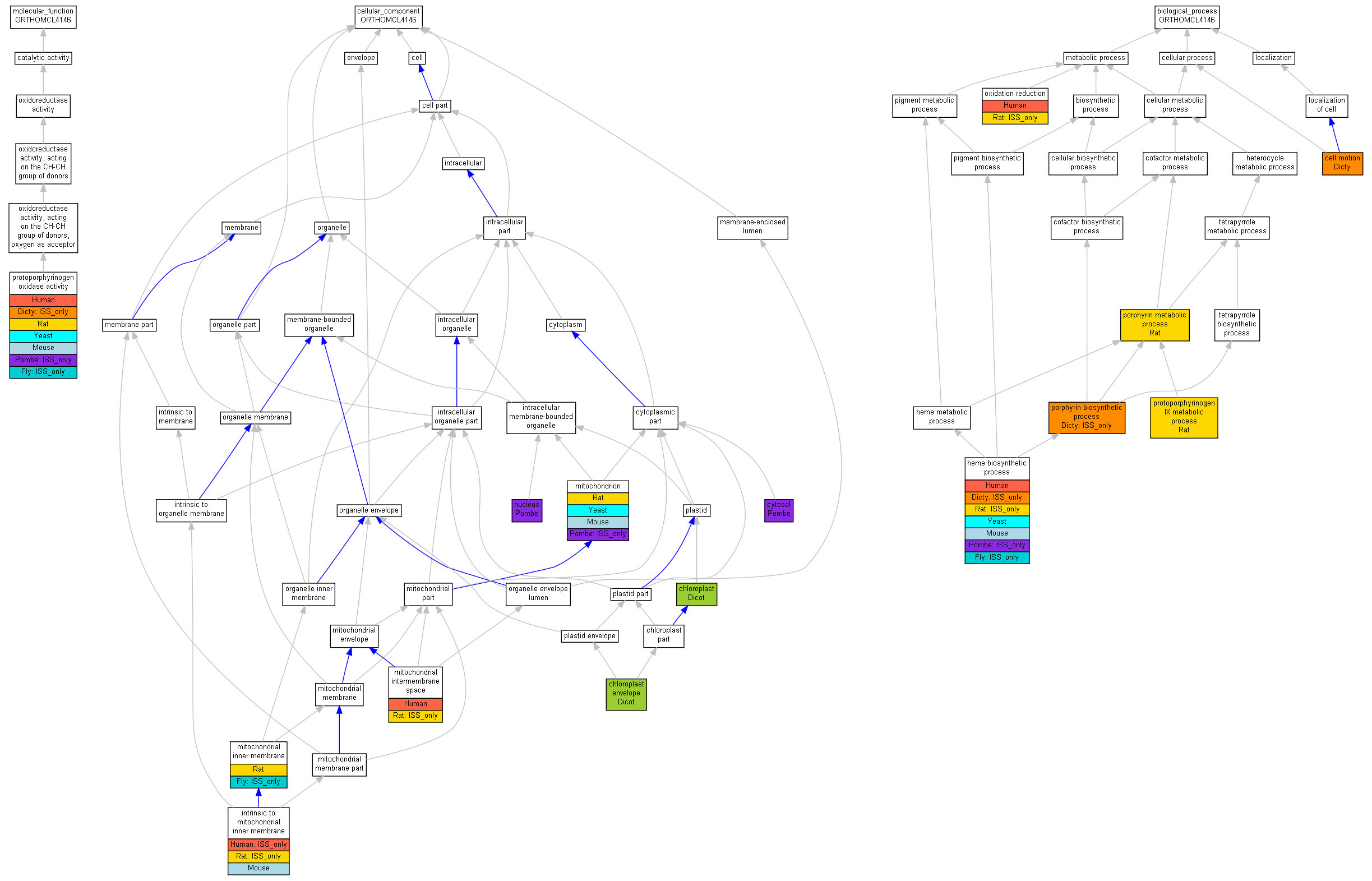
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | X | Zfish | (Fly) | X | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | X | Zfish | (Fly) | X | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | X | Zfish | (Fly) | X | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0031975 | envelope | Human | Mouse | Rat | X | Zfish | (Fly) | X | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0031974 | membrane-enclosed lumen | Human | Mouse | (Rat) | X | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | X | Zfish | (Fly) | X | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | Rat | X | Zfish | (Fly) | X | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | X | Zfish | (Fly) | X | Dicty | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0051179 | localization | Human | Mouse | Rat | X | Zfish | Fly | X | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | X | Zfish | (Fly) | X | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | P50336 | IDA | Human | PMID:7713909 | |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | P50336 | EXP | Human | PMID:8771201 | |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | Ppox | IDA | Mouse | MGI:MGI:3687447|PMID:3346226 | |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | Ppox | IDA | Rat | RGD:1599180|PMID:3663105 | |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | Ppox | ISO | RGD:1321507 | Rat | RGD:1624291 |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | Ppox | ISO | RGD:1321506 | Rat | RGD:1624291 |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | Ppox | ISS | Fly | FB:FBrf0071734 | |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | Ppox | ISS | UniProtKB:P50336 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | Ppox | ISS | UniProtKB:P50336 | Fly | FB:FBrf0159903 |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | hemG | ISS | SGD:S000000816 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | HEM14 | ISS | Yeast | SGD_REF:S000051153|PMID:8948097 | |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | HEM14 | IMP | Yeast | SGD_REF:S000051153|PMID:8948097 | |
| Molecular Function | GO:0004729 | protoporphyrinogen oxidase activity | hem14 | ISS | UniProtKB:P40012 | Pombe | PMID:17072883 |
| Cellular Component | GO:0009507 | chloroplast | PPOX | IDA | Dicot | TAIR:Publication:501712079|PMID:15028209 | |
| Cellular Component | GO:0009507 | chloroplast | PPOX | IDA | Dicot | TAIR:Publication:501712079|PMID:15028209 | |
| Cellular Component | GO:0009507 | chloroplast | PPOX | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0009507 | chloroplast | PPOX | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0009507 | chloroplast | AT5G14220.1 | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0009941 | chloroplast envelope | PPOX | IDA | Dicot | TAIR:Publication:501710686|PMID:12938931 | |
| Cellular Component | GO:0009941 | chloroplast envelope | PPOX | IDA | Dicot | TAIR:Publication:501710686|PMID:12938931 | |
| Cellular Component | GO:0009941 | chloroplast envelope | AT5G14220.1 | IDA | Dicot | TAIR:Publication:501710686|PMID:12938931 | |
| Cellular Component | GO:0005829 | cytosol | hem14 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0031304 | intrinsic to mitochondrial inner membrane | P50336 | ISS | UniProtKB:P51175 | Human | GO_REF:0000024 |
| Cellular Component | GO:0031304 | intrinsic to mitochondrial inner membrane | Ppox | IDA | Mouse | MGI:MGI:3687447|PMID:3346226 | |
| Cellular Component | GO:0031304 | intrinsic to mitochondrial inner membrane | Ppox | ISO | RGD:1321507 | Rat | RGD:1624291 |
| Cellular Component | GO:0005743 | mitochondrial inner membrane | Ppox | IDA | Rat | RGD:1599183|PMID:3996415 | |
| Cellular Component | GO:0005743 | mitochondrial inner membrane | Ppox | ISS | UniProtKB:P50336 | Fly | FB:FBrf0159903 |
| Cellular Component | GO:0005758 | mitochondrial intermembrane space | P50336 | EXP | Human | PMID:8771201 | |
| Cellular Component | GO:0005758 | mitochondrial intermembrane space | Ppox | ISO | RGD:1321506 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | Ppox | IDA | Mouse | MGI:MGI:2682130|PMID:14651853 | |
| Cellular Component | GO:0005739 | mitochondrion | Ppox | IDA | Rat | RGD:1599180|PMID:3663105 | |
| Cellular Component | GO:0005739 | mitochondrion | Ppox | ISO | RGD:1321507 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | HEM14 | IDA | Yeast | SGD_REF:S000075100|PMID:14576278 | |
| Cellular Component | GO:0005739 | mitochondrion | HEM14 | IDA | Yeast | SGD_REF:S000117178|PMID:16823961 | |
| Cellular Component | GO:0005739 | mitochondrion | HEM14 | ISS | Yeast | SGD_REF:S000051153|PMID:8948097 | |
| Cellular Component | GO:0005739 | mitochondrion | hem14 | ISS | UniProtKB:P40012 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005634 | nucleus | hem14 | IDA | Pombe | PMID:16823372 | |
| Biological Process | GO:0006928 | cell motion | hemG | IGI | dictyBase:DDB0214900 | Dicty | dictyBase_REF:11709|PMID:18164290 |
| Biological Process | GO:0006783 | heme biosynthetic process | P50336 | IDA | Human | PMID:7713909 | |
| Biological Process | GO:0006783 | heme biosynthetic process | Ppox | IDA | Mouse | MGI:MGI:3687447|PMID:3346226 | |
| Biological Process | GO:0006783 | heme biosynthetic process | Ppox | ISO | RGD:1321507 | Rat | RGD:1624291 |
| Biological Process | GO:0006783 | heme biosynthetic process | Ppox | ISO | RGD:1321506 | Rat | RGD:1624291 |
| Biological Process | GO:0006783 | heme biosynthetic process | Ppox | ISS | UniProtKB:P50336 | Fly | FB:FBrf0159903 |
| Biological Process | GO:0006783 | heme biosynthetic process | hemG | ISS | SGD:S000000816 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006783 | heme biosynthetic process | HEM14 | IMP | Yeast | SGD_REF:S000051153|PMID:8948097 | |
| Biological Process | GO:0006783 | heme biosynthetic process | hem14 | ISS | UniProtKB:P40012 | Pombe | PMID:17072883 |
| Biological Process | GO:0055114 | oxidation reduction | P50336 | IDA | Human | PMID:7713909 | |
| Biological Process | GO:0055114 | oxidation reduction | Ppox | ISO | RGD:1321506 | Rat | RGD:1624291 |
| Biological Process | GO:0006779 | porphyrin biosynthetic process | hemG | ISS | InterPro:IPR004572 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006778 | porphyrin metabolic process | Ppox | IDA | Rat | RGD:1599180|PMID:3663105 | |
| Biological Process | GO:0046501 | protoporphyrinogen IX metabolic process | Ppox | IDA | Rat | RGD:1599180|PMID:3663105 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |