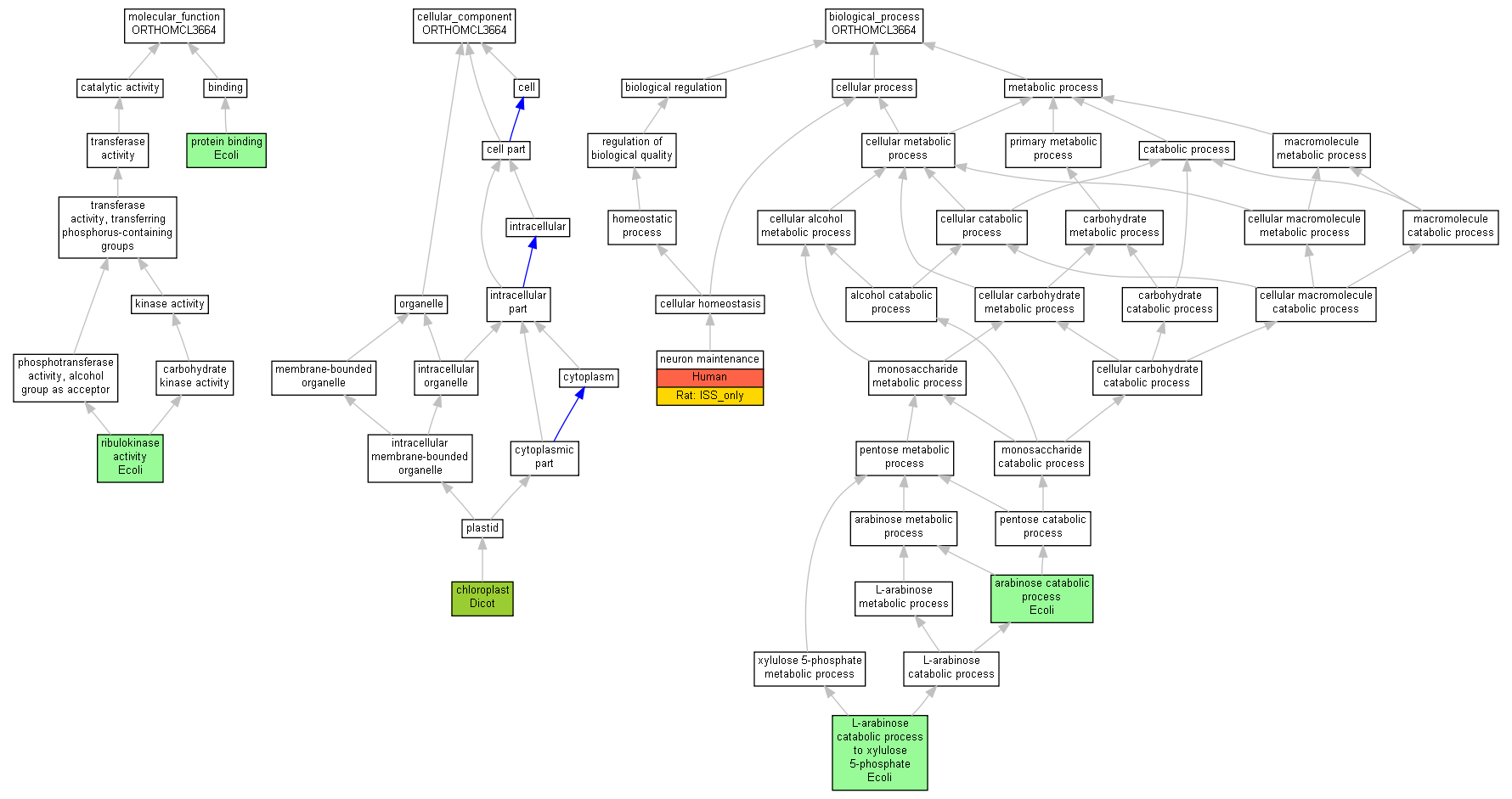
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | X | Fly | X | X | Dicot | Yeast | X | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | X | Fly | X | X | Dicot | Yeast | X | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | X | Fly | X | X | Dicot | Yeast | X | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | X | Fly | X | X | Dicot | Yeast | X | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | X | Fly | X | X | Dicot | Yeast | X | Ecoli |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | (Rat) | Chicken | X | Fly | X | X | Dicot | Yeast | X | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | X | Fly | X | X | Dicot | Yeast | X | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | X | Fly | X | X | Dicot | Yeast | X | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | araB | IPI | UniProtKB:P18843 | Ecoli | PMID:16606699 |
| Molecular Function | GO:0008741 | ribulokinase activity | araB | IDA | Ecoli | PMID:11747300 | |
| Molecular Function | GO:0008741 | ribulokinase activity | araB | IMP | Ecoli | PMID:13829634 | |
| Cellular Component | GO:0009507 | chloroplast | AT4G30310.1 | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0009507 | chloroplast | AT4G30310.2 | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0009507 | chloroplast | AT4G30310.3 | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Biological Process | GO:0019568 | arabinose catabolic process | araB | IMP | Ecoli | PMID:13829634 | |
| Biological Process | GO:0019569 | L-arabinose catabolic process to xylulose 5-phosphate | araB | IEP | Ecoli | PMID:7768852 | |
| Biological Process | GO:0070050 | neuron maintenance | Q96C11 | IMP | Human | PMID:17671248 | |
| Biological Process | GO:0070050 | neuron maintenance | Fggy | ISO | RGD:1605365 | Rat | RGD:1624291 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |