| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | (Rat) | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | (Rat) | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | (Rat) | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | (Rat) | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | (Rat) | Chicken | X | Fly | Worm | X | Dicot | Yeast | Pombe | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
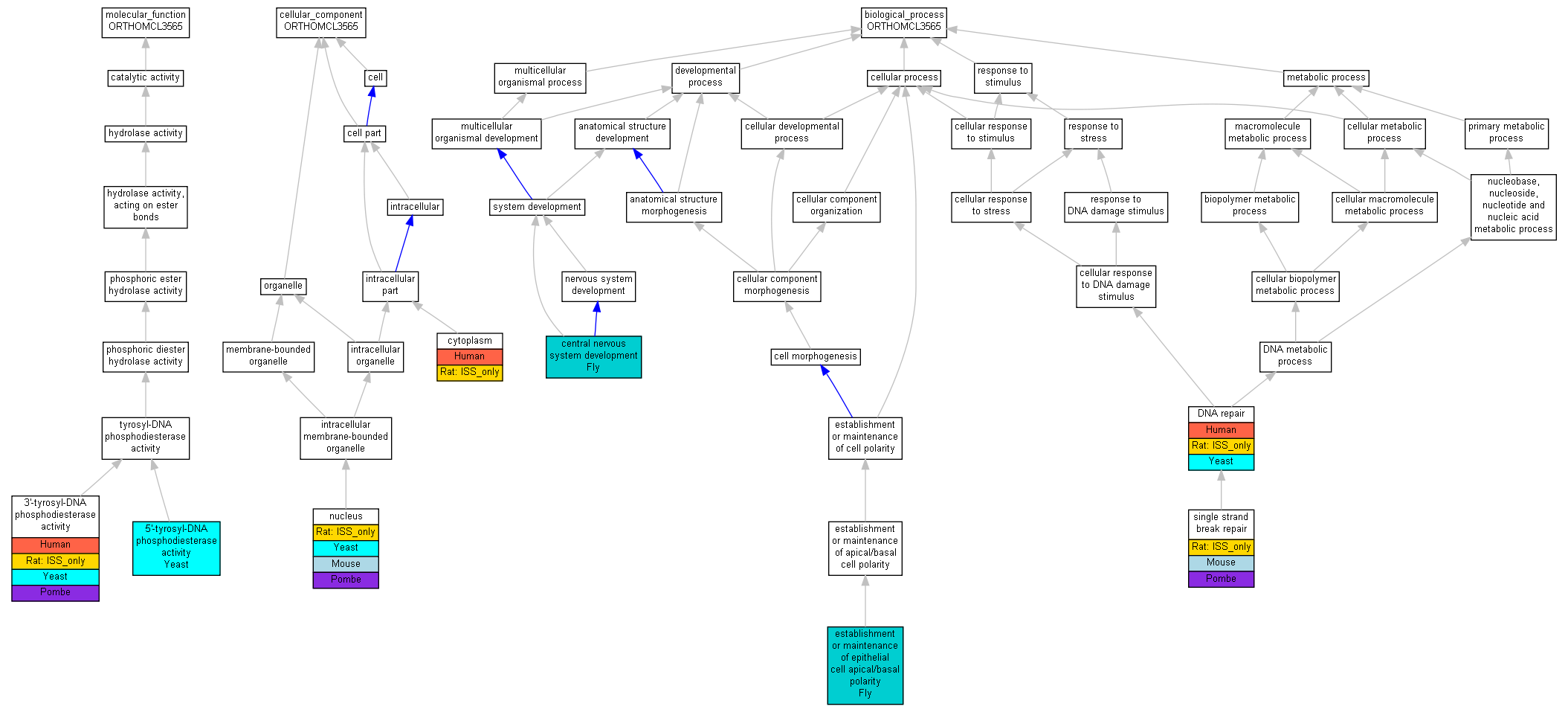
| Molecular Function | GO:0017005 | 3'-tyrosyl-DNA phosphodiesterase activity | Q9NUW8 | IDA | Human | PMID:17948061 | |
| Molecular Function | GO:0017005 | 3'-tyrosyl-DNA phosphodiesterase activity | Tdp1 | ISO | RGD:1320052 | Rat | RGD:1624291 |
| Molecular Function | GO:0017005 | 3'-tyrosyl-DNA phosphodiesterase activity | TDP1 | IDA | Yeast | SGD_REF:S000064819|PMID:10521354 | |
| Molecular Function | GO:0017005 | 3'-tyrosyl-DNA phosphodiesterase activity | tdp1 | IMP | Pombe | PMID:19197239 | |
| Molecular Function | GO:0070260 | 5'-tyrosyl-DNA phosphodiesterase activity | TDP1 | IDA | Yeast | SGD_REF:S000116644|PMID:16751265 | |
| Cellular Component | GO:0005737 | cytoplasm | Q9NUW8 | IDA | Human | PMID:17948061 | |
| Cellular Component | GO:0005737 | cytoplasm | Tdp1 | ISO | RGD:1320052 | Rat | RGD:1624291 |
| Cellular Component | GO:0005634 | nucleus | Tdp1 | IDA | Mouse | MGI:MGI:3763975|PMID:17948061 | |
| Cellular Component | GO:0005634 | nucleus | Tdp1 | ISO | RGD:1617429 | Rat | RGD:1624291 |
| Cellular Component | GO:0005634 | nucleus | TDP1 | IDA | Yeast | SGD_REF:S000071811|PMID:12206772 | |
| Cellular Component | GO:0005634 | nucleus | tdp1 | ISS | SGD:000000427 | Pombe | GOC:unpublished |
| Cellular Component | GO:0005634 | nucleus | tdp1 | IDA | Pombe | PMID:16823372 | |
| Biological Process | GO:0007417 | central nervous system development | gkt | IMP | Fly | FB:FBrf0180073|PMID:15556867 | |
| Biological Process | GO:0006281 | DNA repair | Q9NUW8 | IDA | Human | PMID:17948061 | |
| Biological Process | GO:0006281 | DNA repair | Tdp1 | ISO | RGD:1320052 | Rat | RGD:1624291 |
| Biological Process | GO:0006281 | DNA repair | TDP1 | IMP | Yeast | SGD_REF:S000064819|PMID:10521354 | |
| Biological Process | GO:0006281 | DNA repair | TDP1 | IMP | Yeast | SGD_REF:S000079818|PMID:15135727 | |
| Biological Process | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | gkt | IMP | Fly | FB:FBrf0180073|PMID:15556867 | |
| Biological Process | GO:0000012 | single strand break repair | Tdp1 | IMP | MGI:MGI:3818340 | Mouse | MGI:MGI:3763978|PMID:17914460 |
| Biological Process | GO:0000012 | single strand break repair | Tdp1 | IMP | MGI:MGI:3818340 | Mouse | MGI:MGI:3763975|PMID:17948061 |
| Biological Process | GO:0000012 | single strand break repair | Tdp1 | ISO | RGD:1617429 | Rat | RGD:1624291 |
| Biological Process | GO:0000012 | single strand break repair | tdp1 | IMP | Pombe | PMID:19197239 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |