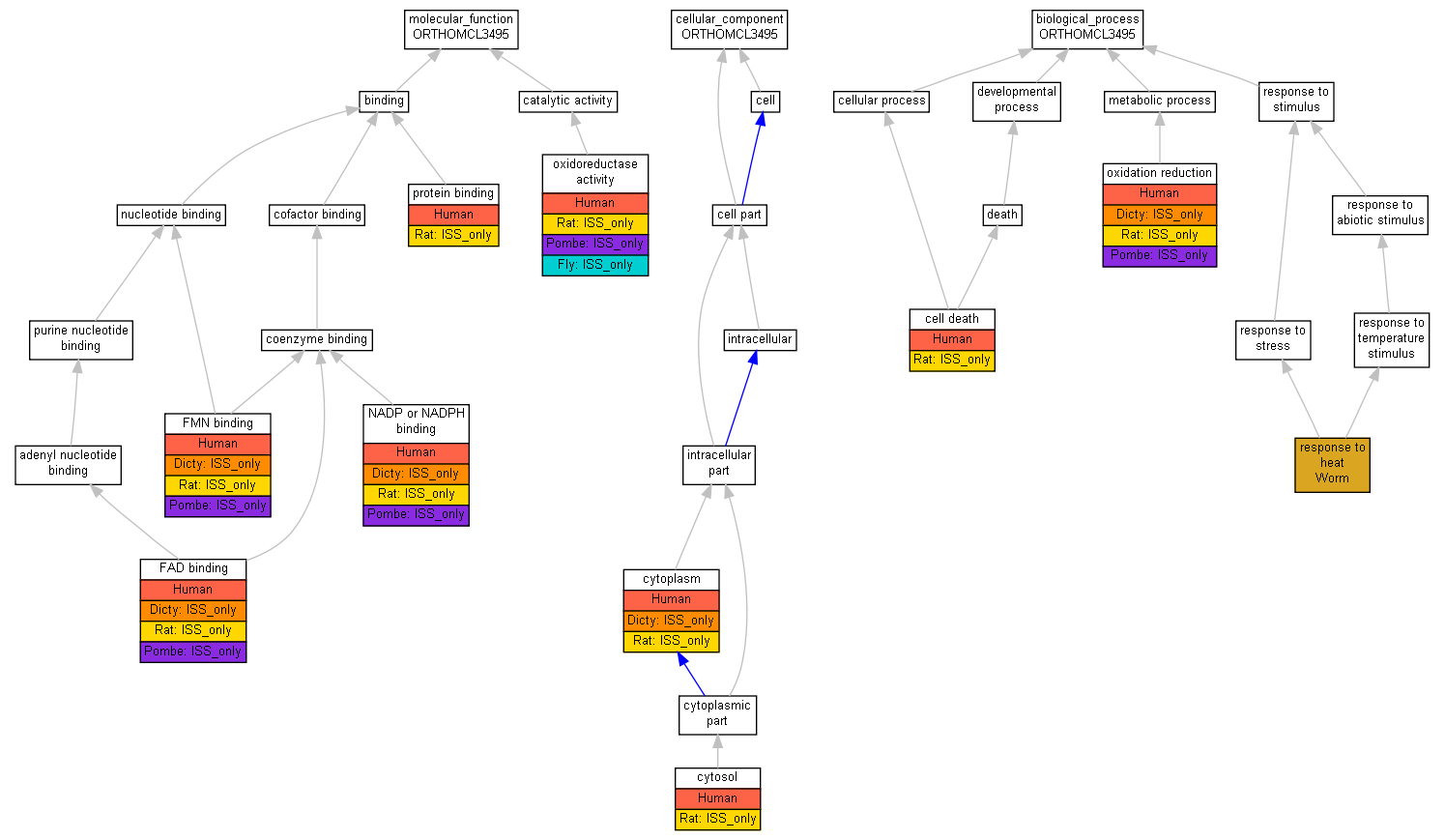
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | (Rat) | X | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | X | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | X | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0050660 | FAD binding | Q9UHB4 | IDA | Human | PMID:10625700 | |
| Molecular Function | GO:0050660 | FAD binding | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Molecular Function | GO:0050660 | FAD binding | redC | ISS | UniProt:Q9UHB4 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0050660 | FAD binding | SPAC1296.06 | ISS | UniProtKB:NDOR1_HUMAN | Pombe | GOC:unpublished |
| Molecular Function | GO:0010181 | FMN binding | Q9UHB4 | IDA | Human | PMID:10625700 | |
| Molecular Function | GO:0010181 | FMN binding | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Molecular Function | GO:0010181 | FMN binding | redC | ISS | UniProt:Q9UHB4 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0010181 | FMN binding | SPAC1296.06 | ISS | UniProtKB:NDOR1_HUMAN | Pombe | GOC:unpublished |
| Molecular Function | GO:0050661 | NADP or NADPH binding | Q9UHB4 | IDA | Human | PMID:15900210 | |
| Molecular Function | GO:0050661 | NADP or NADPH binding | Q9UHB4 | IDA | Human | PMID:10625700 | |
| Molecular Function | GO:0050661 | NADP or NADPH binding | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Molecular Function | GO:0050661 | NADP or NADPH binding | redC | ISS | UniProt:Q9UHB4 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0050661 | NADP or NADPH binding | SPAC1296.06 | ISS | UniProtKB:NDOR1_HUMAN | Pombe | GOC:unpublished |
| Molecular Function | GO:0016491 | oxidoreductase activity | Q9UHB4 | IDA | Human | PMID:10625700 | |
| Molecular Function | GO:0016491 | oxidoreductase activity | Q9UHB4 | IDA | Human | PMID:12631275 | |
| Molecular Function | GO:0016491 | oxidoreductase activity | Q9UHB4 | IDA | Human | PMID:15900210 | |
| Molecular Function | GO:0016491 | oxidoreductase activity | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Molecular Function | GO:0016491 | oxidoreductase activity | CG13667 | ISS | EMBL:AY077845 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0016491 | oxidoreductase activity | SPAC1296.06 | ISS | UniProtKB:NDOR1_HUMAN | Pombe | GOC:unpublished |
| Molecular Function | GO:0005515 | protein binding | Q9UHB4 | IPI | UniProtKB:Q96C86 | Human | PMID:16140270 |
| Molecular Function | GO:0005515 | protein binding | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | Q9UHB4 | IDA | Human | PMID:10625700 | |
| Cellular Component | GO:0005737 | cytoplasm | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | redC | ISS | UniProt:Q9UHB4 | Dicty | dictyBase_REF:10155 |
| Cellular Component | GO:0005829 | cytosol | Q9UHB4 | IDA | Human | PMID:16140270 | |
| Cellular Component | GO:0005829 | cytosol | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Biological Process | GO:0008219 | cell death | Q9UHB4 | IDA | Human | PMID:16140270 | |
| Biological Process | GO:0008219 | cell death | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Biological Process | GO:0055114 | oxidation reduction | Q9UHB4 | IDA | Human | PMID:10625700 | |
| Biological Process | GO:0055114 | oxidation reduction | Q9UHB4 | IDA | Human | PMID:12631275 | |
| Biological Process | GO:0055114 | oxidation reduction | Q9UHB4 | IDA | Human | PMID:15900210 | |
| Biological Process | GO:0055114 | oxidation reduction | Ndor1 | ISO | RGD:1318271 | Rat | RGD:1624291 |
| Biological Process | GO:0055114 | oxidation reduction | redC | ISS | UniProt:Q9UHB4 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0055114 | oxidation reduction | SPAC1296.06 | ISS | UniProtKB:NDOR1_HUMAN | Pombe | GOC:unpublished |
| Biological Process | GO:0009408 | response to heat | fre-1 | IEP | Worm | WB:WBPaper00006128 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |