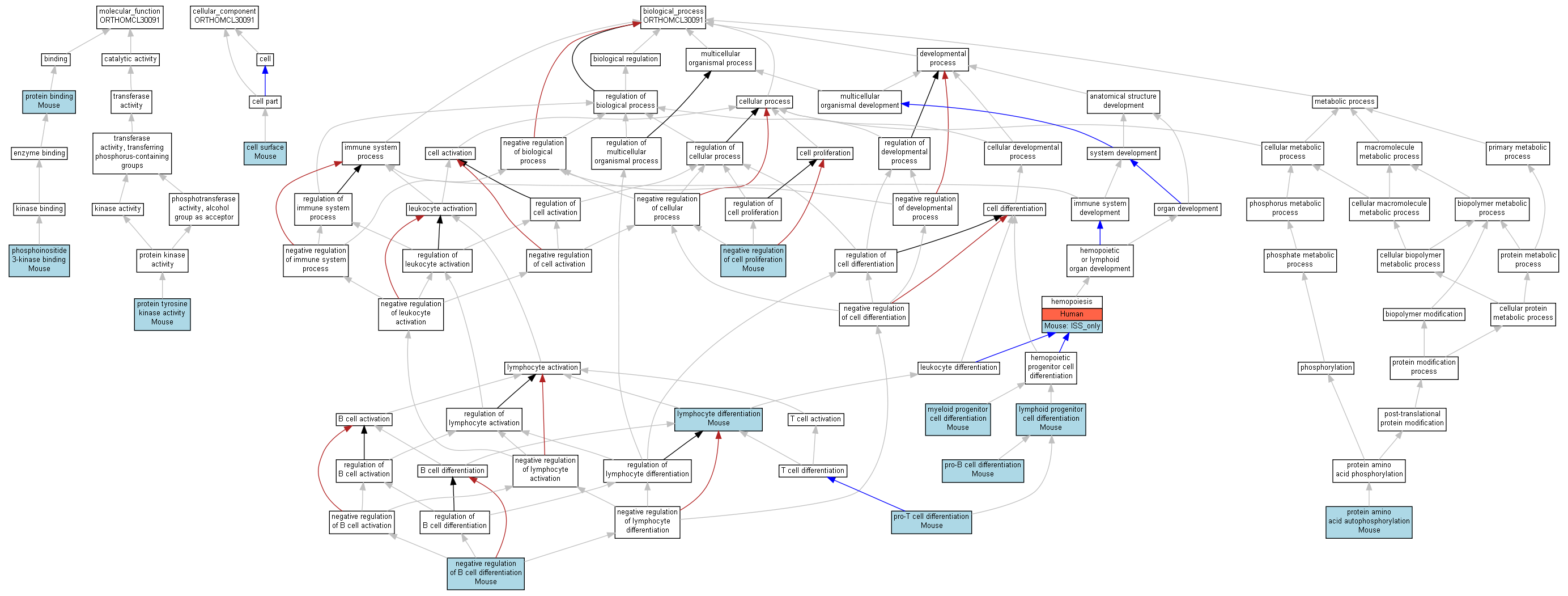
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Biological Process | GO:0002376 | immune system process | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Biological Process | GO:0048519 | negative regulation of biological process | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | X | X | X | X | X | X | X | X | X | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0043548 | phosphoinositide 3-kinase binding | Flt3 | IDA | Mouse | MGI:MGI:67015|PMID:8183574 | |
| Molecular Function | GO:0005515 | protein binding | Flt3 | IPI | UniProtKB:Q91W14 | Mouse | MGI:MGI:1340869|PMID:10409713 |
| Molecular Function | GO:0005515 | protein binding | Flt3 | IPI | UniProtKB:P49772 | Mouse | MGI:MGI:83693|PMID:8896402 |
| Molecular Function | GO:0005515 | protein binding | Flt3 | IPI | UniProtKB:P23727 | Mouse | MGI:MGI:67015|PMID:8183574 |
| Molecular Function | GO:0005515 | protein binding | Flt3 | IPI | UniProtKB:P49772 | Mouse | MGI:MGI:71400|PMID:7531700 |
| Molecular Function | GO:0005515 | protein binding | Flt3 | IPI | UniProtKB:P49772 | Mouse | MGI:MGI:64536|PMID:7505204 |
| Molecular Function | GO:0004713 | protein tyrosine kinase activity | Flt3 | IDA | Mouse | MGI:MGI:3814071|PMID:8384358 | |
| Cellular Component | GO:0009986 | cell surface | Flt3 | IDA | Mouse | MGI:MGI:1206210|PMID:9620281 | |
| Cellular Component | GO:0009986 | cell surface | Flt3 | IDA | Mouse | MGI:MGI:75228|PMID:7630197 | |
| Biological Process | GO:0030097 | hemopoiesis | P36888 | IDA | Human | PMID:7507245 | |
| Biological Process | GO:0030097 | hemopoiesis | Flt3 | ISO | UniProtKB:P36888 | Mouse | MGI:MGI:64561|PMID:7507245 |
| Biological Process | GO:0030098 | lymphocyte differentiation | Flt3 | IDA | Mouse | MGI:MGI:2388504|PMID:12387740 | |
| Biological Process | GO:0002320 | lymphoid progenitor cell differentiation | Flt3 | IGI | MGI:MGI:96677 | Mouse | MGI:MGI:74655|PMID:7621074 |
| Biological Process | GO:0002318 | myeloid progenitor cell differentiation | Flt3 | IGI | MGI:MGI:96677 | Mouse | MGI:MGI:74655|PMID:7621074 |
| Biological Process | GO:0002318 | myeloid progenitor cell differentiation | Flt3 | IMP | MGI:MGI:3783339|MGI:MGI:2176073 | Mouse | MGI:MGI:3778743|PMID:18245664 |
| Biological Process | GO:0045578 | negative regulation of B cell differentiation | Flt3 | IDA | Mouse | MGI:MGI:3623701|PMID:16618805 | |
| Biological Process | GO:0008285 | negative regulation of cell proliferation | Flt3 | IMP | Mouse | MGI:MGI:69294|PMID:7919361 | |
| Biological Process | GO:0002328 | pro-B cell differentiation | Flt3 | IMP | MGI:MGI:2384051 | Mouse | MGI:MGI:74655|PMID:7621074 |
| Biological Process | GO:0002572 | pro-T cell differentiation | Flt3 | IMP | MGI:MGI:2384051 | Mouse | MGI:MGI:74655|PMID:7621074 |
| Biological Process | GO:0046777 | protein amino acid autophosphorylation | Flt3 | IDA | Mouse | MGI:MGI:3814071|PMID:8384358 | |
| Biological Process | GO:0046777 | protein amino acid autophosphorylation | Flt3 | IDA | Mouse | MGI:MGI:67015|PMID:8183574 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |