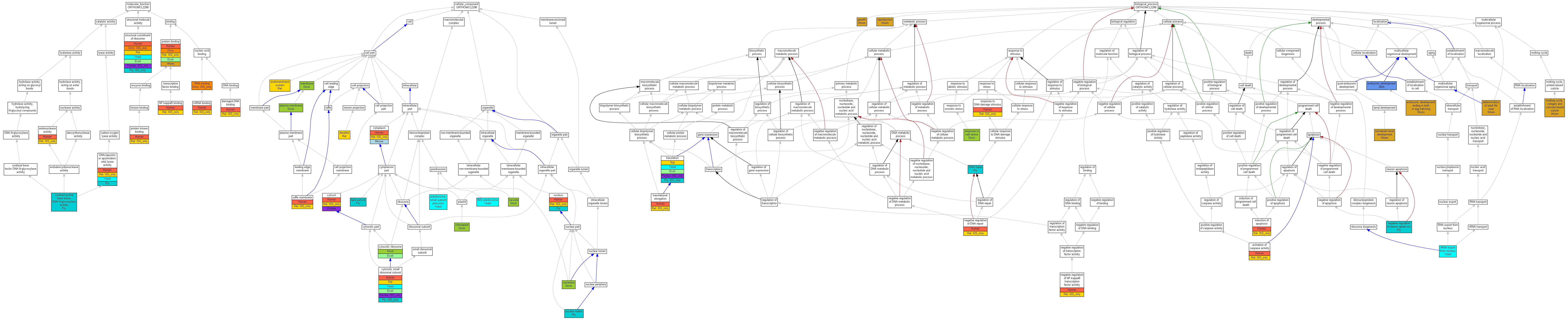
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Molecular Function | GO:0005198 | structural molecule activity | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0032991 | macromolecular complex | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0031974 | membrane-enclosed lumen | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0051234 | establishment of localization | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0051179 | localization | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0048519 | negative regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0000003 | reproduction | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003684 | damaged DNA binding | P23396 | IDA | Human | PMID:16737853 | |
| Molecular Function | GO:0003684 | damaged DNA binding | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Molecular Function | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity | P23396 | IDA | Human | PMID:16737853 | |
| Molecular Function | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Molecular Function | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity | RpS3 | IDA | Fly | FB:FBrf0076708|PMID:7929231 | |
| Molecular Function | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity | RpS3 | IDA | Fly | FB:FBrf0087792|PMID:8641296 | |
| Molecular Function | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity | RPS3 | IDA | Yeast | SGD_REF:S000069743|PMID:11561734 | |
| Molecular Function | GO:0004519 | endonuclease activity | P23396 | IMP | Human | PMID:16584637 | |
| Molecular Function | GO:0004519 | endonuclease activity | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Molecular Function | GO:0003729 | mRNA binding | P23396 | IDA | Human | PMID:18464793 | |
| Molecular Function | GO:0003729 | mRNA binding | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Molecular Function | GO:0051059 | NF-kappaB binding | P23396 | IPI | UniProtKB:Q04206 | Human | PMID:18045535 |
| Molecular Function | GO:0051059 | NF-kappaB binding | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Molecular Function | GO:0008534 | oxidized purine base lesion DNA N-glycosylase activity | RpS3 | IDA | Fly | FB:FBrf0087792|PMID:8641296 | |
| Molecular Function | GO:0005515 | protein binding | P23396 | IPI | UniProtKB:O15527 | Human | PMID:15518571 |
| Molecular Function | GO:0005515 | protein binding | P23396 | IPI | UniProtKB:P27695 | Human | PMID:15518571 |
| Molecular Function | GO:0005515 | protein binding | P23396 | IPI | UniProtKB:P15531 | Human | PMID:16814409 |
| Molecular Function | GO:0005515 | protein binding | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Molecular Function | GO:0005515 | protein binding | rps-3 | IPI | UniProtKB:O02327 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | rps3 | IPI | dictyBase:DDB0001984 | Dicty | dictyBase_REF:11031|PMID:15653432 |
| Molecular Function | GO:0005515 | protein binding | rpsC | IPI | UniProtKB:P16528 | Ecoli | PMID:16606699 |
| Molecular Function | GO:0019901 | protein kinase binding | P23396 | IPI | UniProtKB:P27361 | Human | PMID:15950189 |
| Molecular Function | GO:0019901 | protein kinase binding | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Molecular Function | GO:0003723 | RNA binding | rps3 | ISS | UniProt:P23396 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | P23396 | IDA | Human | PMID:15883184 | |
| Molecular Function | GO:0003735 | structural constituent of ribosome | Rps3 | IC | GO:0022627 | Rat | RGD:2299084|PMID:12876473 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RpS3 | ISS | UniProtKB:P17073 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | rps3 | ISS | UniProt:P23396 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RPS3 | IDA | Yeast | SGD_REF:S000127723|PMID:18782943 | |
| Molecular Function | GO:0003735 | structural constituent of ribosome | rps3 | ISS | SGD:S000005122 | Pombe | PMID:17072883 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | rpsC | IDA | Ecoli | PMID:4587209 | |
| Cellular Component | GO:0030686 | 90S preribosome | RPS3 | IDA | Yeast | SGD_REF:S000086600|PMID:12150911 | |
| Cellular Component | GO:0009507 | chloroplast | AT2G31610.1 | IDA | Dicot | TAIR:Publication:501712079|PMID:15028209 | |
| Cellular Component | GO:0009507 | chloroplast | AT2G31610.1 | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0005737 | cytoplasm | P23396 | IDA | Human | PMID:14988002 | |
| Cellular Component | GO:0005737 | cytoplasm | Rps3 | IDA | Mouse | MGI:MGI:2450792|PMID:12388085 | |
| Cellular Component | GO:0005737 | cytoplasm | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | Rps3 | ISO | RGD:736569 | Rat | RGD:1624291 |
| Cellular Component | GO:0005829 | cytosol | P23396 | EXP | Human | PMID:12588972 | |
| Cellular Component | GO:0005829 | cytosol | P23396 | IDA | Human | PMID:16814409 | |
| Cellular Component | GO:0005829 | cytosol | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Cellular Component | GO:0005829 | cytosol | rps3 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | AT3G53870.1 | IDA | Dicot | TAIR:Publication:501714926|PMID:15734919 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | AT2G31610.1 | IDA | Dicot | TAIR:Publication:501714975|PMID:15821981 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | AT3G53870.1 | IDA | Dicot | TAIR:Publication:501714975|PMID:15821981 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | AT5G35530.1 | IDA | Dicot | TAIR:Publication:501714975|PMID:15821981 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | rpsC | IDA | Ecoli | PMID:4587209 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | P23396 | IDA | Human | PMID:15883184 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | P23396 | IDA | Human | PMID:8706699 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | Rps3 | IDA | Rat | RGD:2299084|PMID:12876473 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | Rps3 | IDA | Rat | RGD:2300010|PMID:947902 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | Rps3 | IDA | Rat | RGD:1599573|PMID:2328735 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RpS3 | ISS | UniProtKB:P17073 | Fly | FB:FBrf0105495 |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RPS3 | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RPS3 | IDA | Yeast | SGD_REF:S000127723|PMID:18782943 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RPS3 | IDA | Yeast | SGD_REF:S000127501|PMID:385049 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | rps3 | ISS | SGD:S000005122 | Pombe | PMID:17072883 |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | rpsC | IDA | Ecoli | PMID:4587209 | |
| Cellular Component | GO:0030425 | dendrite | Rps3 | IDA | Rat | RGD:2299084|PMID:12876473 | |
| Cellular Component | GO:0012505 | endomembrane system | Rps3 | IDA | Rat | RGD:2299084|PMID:12876473 | |
| Cellular Component | GO:0005811 | lipid particle | RpS3 | IDA | Fly | FB:FBrf0192764|PMID:16979555 | |
| Cellular Component | GO:0016020 | membrane | AT2G31610.1 | IDA | Dicot | TAIR:Publication:501721401|PMID:17432890 | |
| Cellular Component | GO:0016020 | membrane | AT3G53870.1 | IDA | Dicot | TAIR:Publication:501721401|PMID:17432890 | |
| Cellular Component | GO:0016020 | membrane | AT5G35530.1 | IDA | Dicot | TAIR:Publication:501721401|PMID:17432890 | |
| Cellular Component | GO:0016363 | nuclear matrix | RpS3 | IDA | Fly | FB:FBrf0076708|PMID:7929231 | |
| Cellular Component | GO:0005730 | nucleolus | AT5G35530.1 | IDA | Dicot | TAIR:Publication:501714224|PMID:15496452 | |
| Cellular Component | GO:0005634 | nucleus | P23396 | IDA | Human | PMID:18045535 | |
| Cellular Component | GO:0005634 | nucleus | P23396 | IDA | Human | PMID:14988002 | |
| Cellular Component | GO:0005634 | nucleus | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Cellular Component | GO:0005634 | nucleus | RpS3 | IDA | Fly | FB:FBrf0076708|PMID:7929231 | |
| Cellular Component | GO:0005886 | plasma membrane | AT2G31610.1 | IDA | Dicot | TAIR:Publication:501720626|PMID:17151019 | |
| Cellular Component | GO:0030688 | preribosome, small subunit precursor | RPS3 | IDA | Yeast | SGD_REF:S000116235|PMID:16738661 | |
| Cellular Component | GO:0032587 | ruffle membrane | P23396 | IDA | Human | PMID:16814409 | |
| Cellular Component | GO:0032587 | ruffle membrane | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Cellular Component | GO:0005773 | vacuole | AT2G31610.1 | IDA | Dicot | TAIR:Publication:501714874|PMID:15539469 | |
| Biological Process | GO:0006919 | activation of caspase activity | P23396 | IDA | Human | PMID:14988002 | |
| Biological Process | GO:0006919 | activation of caspase activity | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Biological Process | GO:0008340 | determination of adult life span | rps-3 | IMP | WB:WBRNAi00066262|WB:WBPhenotype:0000039 | Worm | WB:WBPaper00029254|PMID:17411345 |
| Biological Process | GO:0006281 | DNA repair | RpS3 | IGI | EchoBASE:EB0325 | Fly | FB:FBrf0087792|PMID:8641296 |
| Biological Process | GO:0006281 | DNA repair | RpS3 | IGI | EchoBASE:EB1066|EchoBASE:EB0645 | Fly | FB:FBrf0087792|PMID:8641296 |
| Biological Process | GO:0009790 | embryonic development | rps3 | IMP | ZFIN:ZDB-MRPHLNO-070326-1 | Zfish | ZFIN:ZDB-PUB-061229-22|PMID:17183665 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | rps-3 | IMP | WB:WBRNAi00002450|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00004403|PMID:11099034 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | rps-3 | IMP | WB:WBRNAi00002803|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00004403|PMID:11099034 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | rps-3 | IMP | WB:WBRNAi00041007|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00025054|PMID:15791247 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | rps-3 | IMP | WB:WBRNAi00041008|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00025054|PMID:15791247 |
| Biological Process | GO:0040007 | growth | rps-3 | IMP | WB:WBRNAi00066262|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00029254|PMID:17411345 |
| Biological Process | GO:0006917 | induction of apoptosis | P23396 | IDA | Human | PMID:14988002 | |
| Biological Process | GO:0006917 | induction of apoptosis | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Biological Process | GO:0018996 | molting cycle, collagen and cuticulin-based cuticle | rps-3 | IMP | WB:WBRNAi00066859|WB:WBPhenotype:0000638 | Worm | WB:WBPaper00026763|PMID:16122351 |
| Biological Process | GO:0045738 | negative regulation of DNA repair | P23396 | IMP | Human | PMID:17049931 | |
| Biological Process | GO:0045738 | negative regulation of DNA repair | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Biological Process | GO:0043524 | negative regulation of neuron apoptosis | RpS3 | IMP | Fly | FB:FBrf0190282|PMID:16291791 | |
| Biological Process | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | P23396 | IMP | Human | PMID:18045535 | |
| Biological Process | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Biological Process | GO:0002119 | nematode larval development | rps-3 | IMP | WB:WBRNAi00033268|WB:WBPhenotype:0000054 | Worm | WB:WBPaper00024497|PMID:15489339 |
| Biological Process | GO:0002119 | nematode larval development | rps-3 | IMP | WB:WBRNAi00066262|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00029254|PMID:17411345 |
| Biological Process | GO:0000003 | reproduction | rps-3 | IMP | WB:WBRNAi00008425|WB:WBPhenotype:0000689 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0000003 | reproduction | rps-3 | IMP | WB:WBRNAi00024597|WB:WBPhenotype:0000689 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0006974 | response to DNA damage stimulus | P23396 | IEP | Human | PMID:17560175 | |
| Biological Process | GO:0006974 | response to DNA damage stimulus | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| Biological Process | GO:0009651 | response to salt stress | AT2G31610.1 | IEP | Dicot | TAIR:Publication:501723430|PMID:17916636 | |
| Biological Process | GO:0009651 | response to salt stress | AT3G53870.1 | IEP | Dicot | TAIR:Publication:501723430|PMID:17916636 | |
| Biological Process | GO:0009651 | response to salt stress | AT5G35530.1 | IEP | Dicot | TAIR:Publication:501723430|PMID:17916636 | |
| Biological Process | GO:0006407 | rRNA export from nucleus | RPS3 | IMP | Yeast | SGD_REF:S000087225|PMID:16246728 | |
| Biological Process | GO:0006412 | translation | Rps3 | IC | GO:0012505 | Rat | RGD:2299084|PMID:12876473 |
| Biological Process | GO:0006412 | translation | RpS3 | ISS | UniProtKB:P17073 | Fly | FB:FBrf0105495 |
| Biological Process | GO:0006412 | translation | RPS3 | IC | GO:0022627 | Yeast | SGD_REF:S000127723|PMID:18782943 |
| Biological Process | GO:0006412 | translation | RPS3 | IC | GO:0022627 | Yeast | SGD_REF:S000116584|PMID:3533916 |
| Biological Process | GO:0006412 | translation | RPS3 | IC | GO:0022627 | Yeast | SGD_REF:S000127501|PMID:385049 |
| Biological Process | GO:0006412 | translation | rps3 | ISS | SGD:S000005122 | Pombe | PMID:17072883 |
| Biological Process | GO:0006412 | translation | rpsC | IDA | Ecoli | PMID:4579428 | |
| Biological Process | GO:0006414 | translational elongation | P23396 | EXP | Human | PMID:15189156 | |
| Biological Process | GO:0006414 | translational elongation | Rps3 | ISO | RGD:736568 | Rat | RGD:1624291 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |