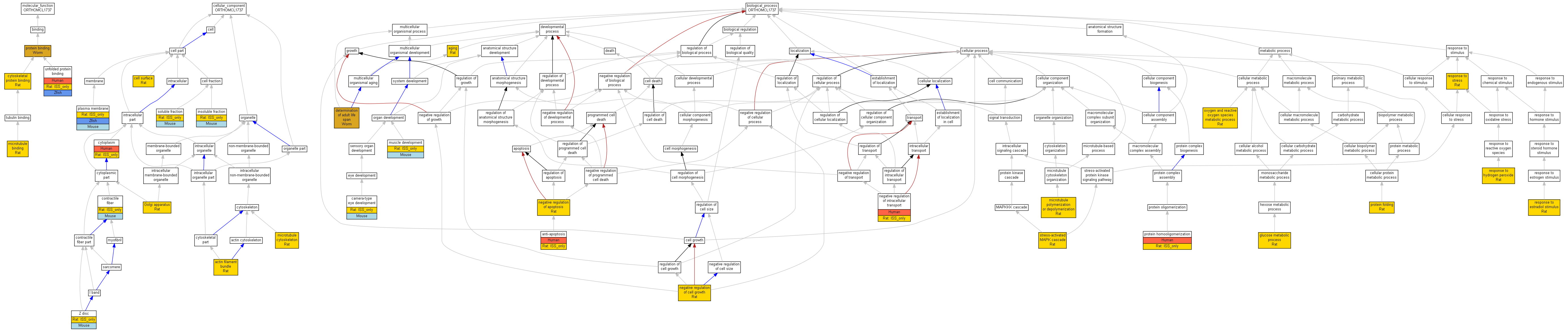
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0010926 | anatomical structure formation | Human | Mouse | (Rat) | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0051234 | establishment of localization | Human | Mouse | (Rat) | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0051179 | localization | Human | Mouse | (Rat) | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | (Rat) | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0048519 | negative regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | X | Worm | X | X | X | X | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008092 | cytoskeletal protein binding | Cryab | IDA | Rat | RGD:1598493|PMID:15075233 | |
| Molecular Function | GO:0008017 | microtubule binding | Cryab | IDA | Rat | RGD:1598493|PMID:15075233 | |
| Molecular Function | GO:0005515 | protein binding | hsp-12.2 | IPI | UniProtKB:Q20165 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | hsp-12.6 | IPI | UniProtKB:P34328 | Worm | PMID:14704431 |
| Molecular Function | GO:0051082 | unfolded protein binding | P02511 | IPI | UniProtKB:P07315 | Human | PMID:16303126 |
| Molecular Function | GO:0051082 | unfolded protein binding | Cryab | ISO | RGD:737518 | Rat | RGD:1624291 |
| Molecular Function | GO:0051082 | unfolded protein binding | cryaba | IDA | Zfish | ZFIN:ZDB-PUB-050209-6|PMID:15692462 | |
| Molecular Function | GO:0051082 | unfolded protein binding | cryabb | IDA | Zfish | ZFIN:ZDB-PUB-060124-23|PMID:16420472 | |
| Cellular Component | GO:0032432 | actin filament bundle | Cryab | IDA | Rat | RGD:1598494|PMID:10625651 | |
| Cellular Component | GO:0009986 | cell surface | Cryab | IDA | Rat | RGD:2303634|PMID:17761354 | |
| Cellular Component | GO:0043292 | contractile fiber | Cryab | IDA | Mouse | MGI:MGI:3028713|PMID:14627610 | |
| Cellular Component | GO:0043292 | contractile fiber | Cryab | ISO | RGD:10400 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | P02511 | IDA | Human | PMID:14752512 | |
| Cellular Component | GO:0005737 | cytoplasm | Cryab | ISO | RGD:737518 | Rat | RGD:1624291 |
| Cellular Component | GO:0005794 | Golgi apparatus | Cryab | IDA | Rat | RGD:1598492|PMID:15339919 | |
| Cellular Component | GO:0005626 | insoluble fraction | Cryab | IDA | Mouse | MGI:MGI:3028713|PMID:14627610 | |
| Cellular Component | GO:0005626 | insoluble fraction | Cryab | ISO | RGD:10400 | Rat | RGD:1624291 |
| Cellular Component | GO:0015630 | microtubule cytoskeleton | Cryab | IDA | Rat | RGD:1598493|PMID:15075233 | |
| Cellular Component | GO:0005886 | plasma membrane | Cryab | IDA | Mouse | MGI:MGI:3028713|PMID:14627610 | |
| Cellular Component | GO:0005886 | plasma membrane | Cryab | ISO | RGD:10400 | Rat | RGD:1624291 |
| Cellular Component | GO:0005886 | plasma membrane | cryaba | IDA | Zfish | ZFIN:ZDB-PUB-080415-15|PMID:18406404 | |
| Cellular Component | GO:0005625 | soluble fraction | Cryab | IDA | Mouse | MGI:MGI:3028713|PMID:14627610 | |
| Cellular Component | GO:0005625 | soluble fraction | Cryab | ISO | RGD:10400 | Rat | RGD:1624291 |
| Cellular Component | GO:0030018 | Z disc | Cryab | IDA | Mouse | MGI:MGI:3028713|PMID:14627610 | |
| Cellular Component | GO:0030018 | Z disc | Cryab | ISO | RGD:10400 | Rat | RGD:1624291 |
| Biological Process | GO:0007568 | aging | Cryab | IEP | Rat | RGD:2303634|PMID:17761354 | |
| Biological Process | GO:0006916 | anti-apoptosis | P02511 | IDA | Human | PMID:14752512 | |
| Biological Process | GO:0006916 | anti-apoptosis | Cryab | ISO | RGD:737518 | Rat | RGD:1624291 |
| Biological Process | GO:0043010 | camera-type eye development | Cryab | IGI | MGI:MGI:88515 | Mouse | MGI:MGI:2682441|PMID:12546709 |
| Biological Process | GO:0043010 | camera-type eye development | Cryab | ISO | RGD:10400 | Rat | RGD:1624291 |
| Biological Process | GO:0008340 | determination of adult life span | hsp-12.6 | IMP | WB:WBRNAi00063155|WB:WBPhenotype:0000039 | Worm | WB:WBPaper00005976|PMID:12845331 |
| Biological Process | GO:0008340 | determination of adult life span | hsp-12.6 | IMP | WB:WBRNAi00063156|WB:WBPhenotype:0000039 | Worm | WB:WBPaper00005976|PMID:12845331 |
| Biological Process | GO:0006006 | glucose metabolic process | Cryab | IMP | Rat | RGD:1598485|PMID:15358243 | |
| Biological Process | GO:0031109 | microtubule polymerization or depolymerization | Cryab | IMP | Rat | RGD:1598493|PMID:15075233 | |
| Biological Process | GO:0007517 | muscle development | Cryab | IGI | MGI:MGI:1916503 | Mouse | MGI:MGI:2154007|PMID:11687538 |
| Biological Process | GO:0007517 | muscle development | Cryab | ISO | RGD:10400 | Rat | RGD:1624291 |
| Biological Process | GO:0043066 | negative regulation of apoptosis | Cryab | IMP | Rat | RGD:1598485|PMID:15358243 | |
| Biological Process | GO:0030308 | negative regulation of cell growth | Cryab | IDA | Rat | RGD:2303630|PMID:18974385 | |
| Biological Process | GO:0032387 | negative regulation of intracellular transport | P02511 | IDA | Human | PMID:14752512 | |
| Biological Process | GO:0032387 | negative regulation of intracellular transport | Cryab | ISO | RGD:737518 | Rat | RGD:1624291 |
| Biological Process | GO:0006800 | oxygen and reactive oxygen species metabolic process | Cryab | IMP | Rat | RGD:1598485|PMID:15358243 | |
| Biological Process | GO:0006457 | protein folding | Cryab | IDA | Rat | RGD:2303633|PMID:18158587 | |
| Biological Process | GO:0051260 | protein homooligomerization | P02511 | IDA | Human | PMID:16303126 | |
| Biological Process | GO:0051260 | protein homooligomerization | Cryab | ISO | RGD:737518 | Rat | RGD:1624291 |
| Biological Process | GO:0032355 | response to estradiol stimulus | Cryab | IEP | Rat | RGD:2303639|PMID:17293487 | |
| Biological Process | GO:0042542 | response to hydrogen peroxide | Cryab | IEP | Rat | RGD:2303631|PMID:18420382 | |
| Biological Process | GO:0006950 | response to stress | Cryab | IPI | Rat | RGD:704398|PMID:11945023 | |
| Biological Process | GO:0051403 | stress-activated MAPK cascade | Cryab | IMP | Rat | RGD:2303631|PMID:18420382 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |