| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | X | X | (Zfish) | Fly | X | X | X | X | X | X |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Molecular Function | GO:0030528 | transcription regulator activity | Human | Mouse | X | X | (Zfish) | Fly | X | X | X | X | X | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Cellular Component | GO:0032991 | macromolecular complex | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | X | X | (Zfish) | Fly | X | X | X | X | X | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | X | X | (Zfish) | Fly | X | X | X | X | X | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | X | X | (Zfish) | Fly | X | X | X | X | X | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0048519 | negative regulation of biological process | Human | Mouse | X | X | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | X | X | (Zfish) | Fly | X | X | X | X | X | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
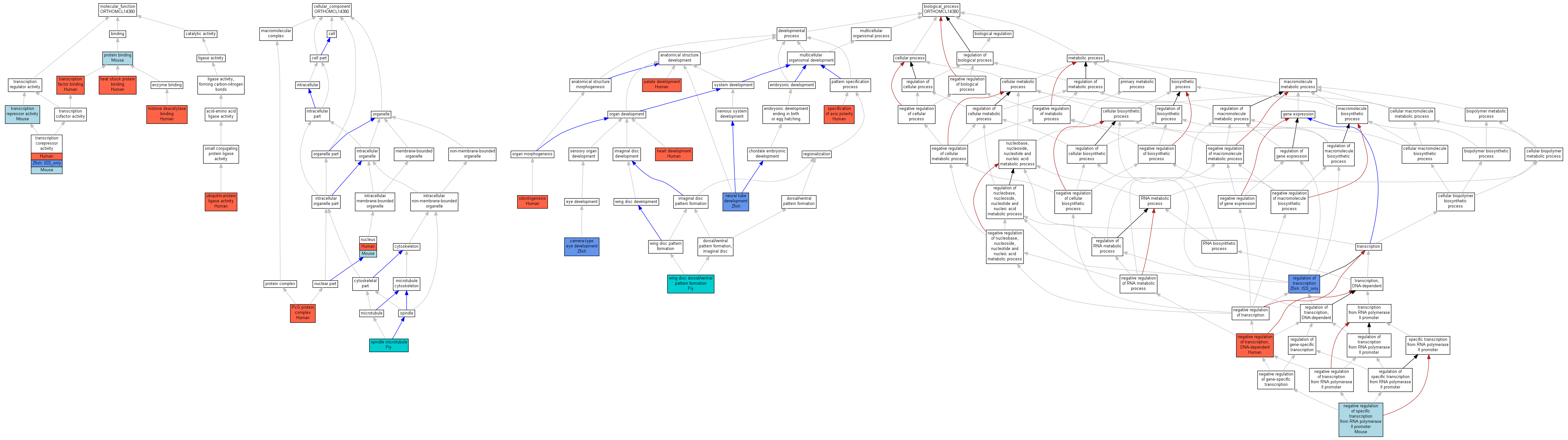
| Molecular Function | GO:0031072 | heat shock protein binding | Q6W2J9 | IDA | Human | PMID:16943429 | |
| Molecular Function | GO:0042826 | histone deacetylase binding | Q6W2J9 | IPI | UniProtKB:Q13547 | Human | PMID:10898795 |
| Molecular Function | GO:0042826 | histone deacetylase binding | Q6W2J9 | IPI | UniProtKB:O15379 | Human | PMID:10898795 |
| Molecular Function | GO:0042826 | histone deacetylase binding | Q6W2J9 | IPI | UniProtKB:Q9UQL6 | Human | PMID:10898795 |
| Molecular Function | GO:0005515 | protein binding | Bcor | IPI | UniProtKB:A2AM29 | Mouse | MGI:MGI:2682643|PMID:12776190 |
| Molecular Function | GO:0003714 | transcription corepressor activity | Q6W2J9 | IDA | Human | PMID:10898795 | |
| Molecular Function | GO:0003714 | transcription corepressor activity | Q6W2J9 | IMP | Human | PMID:15878880 | |
| Molecular Function | GO:0003714 | transcription corepressor activity | Bcor | IPI | UniProtKB:P41183 | Mouse | MGI:MGI:2682643|PMID:12776190 |
| Molecular Function | GO:0003714 | transcription corepressor activity | bcor | ISS | UniProtKB:Q9H233 | Zfish | ZFIN:ZDB-PUB-040312-8|PMID:15004558 |
| Molecular Function | GO:0008134 | transcription factor binding | Q6W2J9 | IPI | UniProtKB:P41182 | Human | PMID:10898795 |
| Molecular Function | GO:0016564 | transcription repressor activity | Bcor | IDA | Mouse | MGI:MGI:2682643|PMID:12776190 | |
| Molecular Function | GO:0004842 | ubiquitin-protein ligase activity | Q6W2J9 | IDA | Human | PMID:16943429 | |
| Cellular Component | GO:0005634 | nucleus | Q6W2J9 | IDA | Human | PMID:10898795 | |
| Cellular Component | GO:0005634 | nucleus | Bcor | IDA | Mouse | MGI:MGI:2682643|PMID:12776190 | |
| Cellular Component | GO:0031519 | PcG protein complex | Q6W2J9 | IDA | Human | PMID:16943429 | |
| Cellular Component | GO:0005876 | spindle microtubule | CG14073 | IDA | Fly | FB:FBrf0182794 | |
| Biological Process | GO:0043010 | camera-type eye development | bcor | IMP | Zfish | ZFIN:ZDB-PUB-040312-8|PMID:15004558 | |
| Biological Process | GO:0007507 | heart development | Q6W2J9 | IMP | Human | PMID:17517692 | |
| Biological Process | GO:0010553 | negative regulation of specific transcription from RNA polymerase II promoter | Bcor | IDA | Mouse | MGI:MGI:2682643|PMID:12776190 | |
| Biological Process | GO:0045892 | negative regulation of transcription, DNA-dependent | Q6W2J9 | IDA | Human | PMID:10898795 | |
| Biological Process | GO:0021915 | neural tube development | bcor | IMP | Zfish | ZFIN:ZDB-PUB-040312-8|PMID:15004558 | |
| Biological Process | GO:0042476 | odontogenesis | Q6W2J9 | IMP | Human | PMID:17517692 | |
| Biological Process | GO:0060021 | palate development | Q6W2J9 | IMP | Human | PMID:17517692 | |
| Biological Process | GO:0045449 | regulation of transcription | bcor | ISS | UniProtKB:Q9H233 | Zfish | ZFIN:ZDB-PUB-040312-8|PMID:15004558 |
| Biological Process | GO:0065001 | specification of axis polarity | Q6W2J9 | IMP | Human | PMID:17517692 | |
| Biological Process | GO:0048190 | wing disc dorsal/ventral pattern formation | CG14073 | IGI | FB:FBgn0000242|FB:FBgn0000099 | Fly | FB:FBrf0202749|PMID:18202376 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |