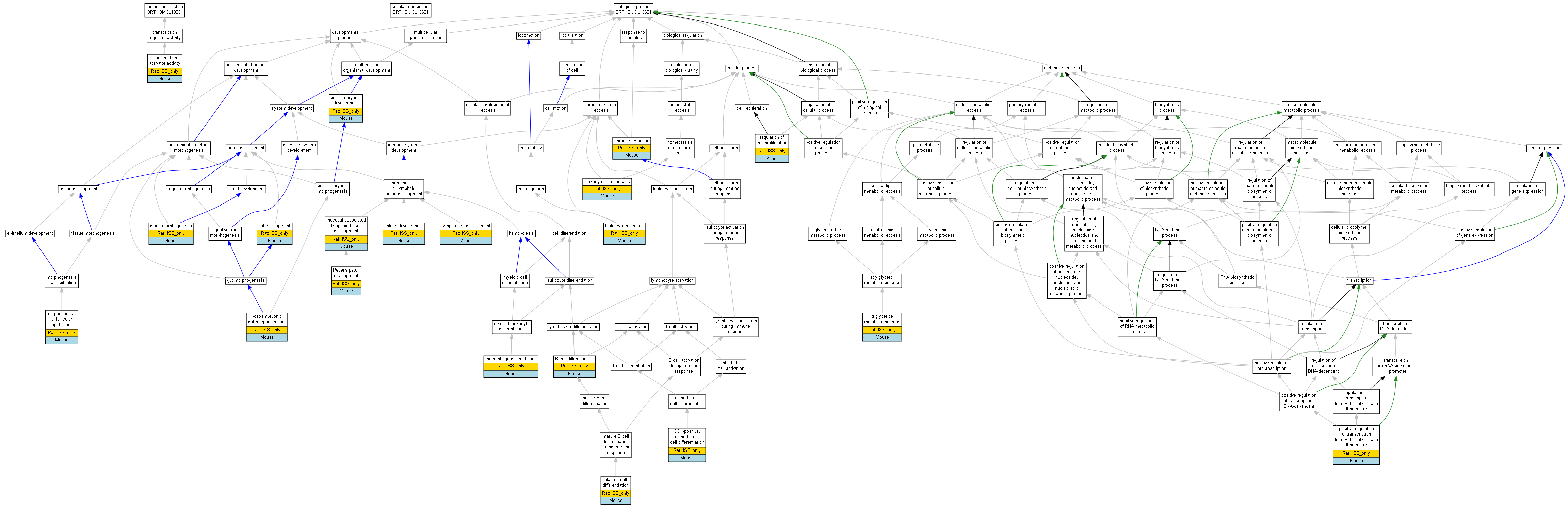
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0030528 | transcription regulator activity | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0002376 | immune system process | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0051179 | localization | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0040011 | locomotion | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | (Rat) | Chicken | Zfish | X | X | X | X | X | X | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016563 | transcription activator activity | Nkx2-3 | IDA | Mouse | MGI:MGI:1858364|PMID:10790368 | |
| Molecular Function | GO:0016563 | transcription activator activity | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0030183 | B cell differentiation | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:2654966|PMID:12682228 |
| Biological Process | GO:0030183 | B cell differentiation | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0043367 | CD4-positive, alpha beta T cell differentiation | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0043367 | CD4-positive, alpha beta T cell differentiation | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0022612 | gland morphogenesis | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0022612 | gland morphogenesis | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0048565 | gut development | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0048565 | gut development | Nkx2-3 | IMP | MGI:MGI:2386680 | Mouse | MGI:MGI:1334782|PMID:10207146 |
| Biological Process | GO:0048565 | gut development | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0006955 | immune response | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:2654966|PMID:12682228 |
| Biological Process | GO:0006955 | immune response | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0001776 | leukocyte homeostasis | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0001776 | leukocyte homeostasis | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0050900 | leukocyte migration | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0050900 | leukocyte migration | Nkx2-3 | IMP | MGI:MGI:2386680 | Mouse | MGI:MGI:1858364|PMID:10790368 |
| Biological Process | GO:0050900 | leukocyte migration | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0048535 | lymph node development | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0048535 | lymph node development | Nkx2-3 | IMP | MGI:MGI:2386680 | Mouse | MGI:MGI:1858364|PMID:10790368 |
| Biological Process | GO:0048535 | lymph node development | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0030225 | macrophage differentiation | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0030225 | macrophage differentiation | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0016333 | morphogenesis of follicular epithelium | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0016333 | morphogenesis of follicular epithelium | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0048537 | mucosal-associated lymphoid tissue development | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0048537 | mucosal-associated lymphoid tissue development | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0048541 | Peyer's patch development | Nkx2-3 | IMP | MGI:MGI:2386680 | Mouse | MGI:MGI:1858364|PMID:10790368 |
| Biological Process | GO:0048541 | Peyer's patch development | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0002317 | plasma cell differentiation | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0002317 | plasma cell differentiation | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | Nkx2-3 | IDA | Mouse | MGI:MGI:1858364|PMID:10790368 | |
| Biological Process | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0009791 | post-embryonic development | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0009791 | post-embryonic development | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0048621 | post-embryonic gut morphogenesis | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0048621 | post-embryonic gut morphogenesis | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0042127 | regulation of cell proliferation | Nkx2-3 | IMP | MGI:MGI:2386680 | Mouse | MGI:MGI:1334782|PMID:10207146 |
| Biological Process | GO:0042127 | regulation of cell proliferation | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0048536 | spleen development | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0048536 | spleen development | Nkx2-3 | IMP | MGI:MGI:2386680 | Mouse | MGI:MGI:1334782|PMID:10207146 |
| Biological Process | GO:0048536 | spleen development | Nkx2-3 | IMP | MGI:MGI:2386680 | Mouse | MGI:MGI:1858364|PMID:10790368 |
| Biological Process | GO:0048536 | spleen development | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| Biological Process | GO:0006641 | triglyceride metabolic process | Nkx2-3 | IMP | MGI:MGI:2448069 | Mouse | MGI:MGI:1888604|PMID:10926756 |
| Biological Process | GO:0006641 | triglyceride metabolic process | Nkx2-3 | ISO | RGD:1318341 | Rat | RGD:1624291 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |