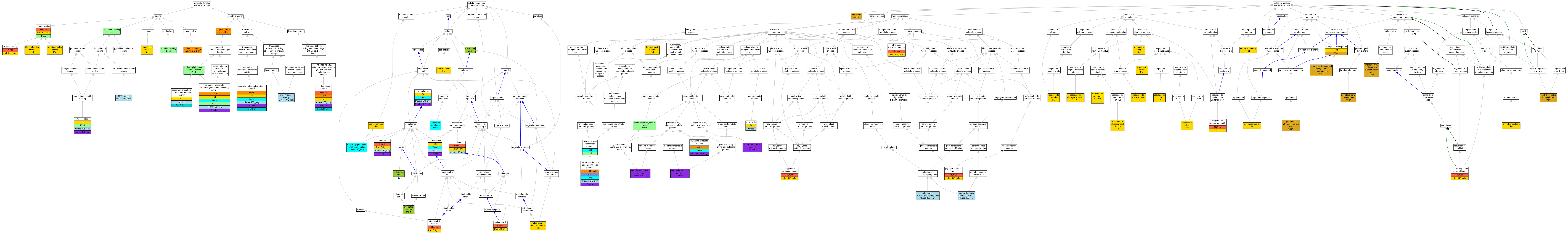
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | (Mouse) | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0031975 | envelope | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0032991 | macromolecular complex | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | (Yeast) | Pombe | Ecoli |
| Cellular Component | GO:0031974 | membrane-enclosed lumen | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0040011 | locomotion | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0051704 | multi-organism process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0000003 | reproduction | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0022414 | reproductive process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016597 | amino acid binding | pyr1-3 | ISS | InterPro:IPR006131 | Dicty | dictyBase_REF:1660|PMID:2917570 |
| Molecular Function | GO:0004070 | aspartate carbamoyltransferase activity | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Molecular Function | GO:0004070 | aspartate carbamoyltransferase activity | Cad | IDA | Rat | RGD:2303536|PMID:1148171 | |
| Molecular Function | GO:0004070 | aspartate carbamoyltransferase activity | r | ISS | NCBI_gi:4757896 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004070 | aspartate carbamoyltransferase activity | pyr1-3 | IDA | Dicty | dictyBase_REF:6651|PMID:2568162 | |
| Molecular Function | GO:0004070 | aspartate carbamoyltransferase activity | URA2 | IDA | Yeast | SGD_REF:S000050154|PMID:10446140 | |
| Molecular Function | GO:0004070 | aspartate carbamoyltransferase activity | URA2 | IMP | Yeast | SGD_REF:S000124960|PMID:4550660 | |
| Molecular Function | GO:0004070 | aspartate carbamoyltransferase activity | ura1 | IGI | UniProtKB:Q09794 | Pombe | PMID:8590465 |
| Molecular Function | GO:0005524 | ATP binding | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Molecular Function | GO:0005524 | ATP binding | Cps1 | IDA | Rat | RGD:2303524|PMID:11577071 | |
| Molecular Function | GO:0005524 | ATP binding | arg4 | ISS | InterPro:IPR005479 | Pombe | PMID:17072883 |
| Molecular Function | GO:0005524 | ATP binding | carB | IMP | Ecoli | PMID:14718657 | |
| Molecular Function | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity | Cps1 | IMP | MGI:MGI:2386969 | Mouse | MGI:MGI:1328875|PMID:9862865 |
| Molecular Function | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity | Cps1 | IDA | Rat | RGD:2303524|PMID:11577071 | |
| Molecular Function | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity | Cps1 | IDA | Rat | RGD:2300098|PMID:4062872 | |
| Molecular Function | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity | Cps1 | ISO | RGD:10389 | Rat | RGD:1624291 |
| Molecular Function | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity | Cps1 | IDA | Rat | RGD:1582379|PMID:3680220 | |
| Molecular Function | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity | r | ISS | NCBI_gi:4757896 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | Cad | IDA | Rat | RGD:2303536|PMID:1148171 | |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | pyr1-3 | IDA | Dicty | dictyBase_REF:6651|PMID:2568162 | |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | CPA2 | IMP | Yeast | SGD_REF:S000057270|PMID:6358221 | |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | CPA2 | ISS | Yeast | SGD_REF:S000057270|PMID:6358221 | |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | URA2 | IDA | Yeast | SGD_REF:S000065636|PMID:5776390 | |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | ura1 | IGI | UniProtKB:Q09794 | Pombe | PMID:8590465 |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | arg4 | ISS | UniProtKB:P03965 | Pombe | PMID:17072883 |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | carB | IGI | EcoliWiki:carA | Ecoli | PMID:1737023 |
| Molecular Function | GO:0004086 | carbamoyl-phosphate synthase activity | carB | IDA | Ecoli | PMID:14718657 | |
| Molecular Function | GO:0004086 | carbamoyl-phosphate synthase activity | carB | IDA | Ecoli | PMID:3549732 | |
| Molecular Function | GO:0004151 | dihydroorotase activity | P27708 | EXP | Human | PMID:8619816 | |
| Molecular Function | GO:0004151 | dihydroorotase activity | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Molecular Function | GO:0004151 | dihydroorotase activity | Cad | IDA | Rat | RGD:2303536|PMID:1148171 | |
| Molecular Function | GO:0004151 | dihydroorotase activity | Cad | ISO | RGD:1348167 | Rat | RGD:1624291 |
| Molecular Function | GO:0004151 | dihydroorotase activity | r | ISS | NCBI_gi:4757896 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004151 | dihydroorotase activity | pyr1-3 | IDA | Dicty | dictyBase_REF:6651|PMID:2568162 | |
| Molecular Function | GO:0004151 | dihydroorotase activity | ura1 | IGI | UniProtKB:Q09794 | Pombe | PMID:8590465 |
| Molecular Function | GO:0019899 | enzyme binding | P27708 | IPI | UniProtKB:Q99638 | Human | PMID:15326225 |
| Molecular Function | GO:0019899 | enzyme binding | Cad | ISO | RGD:1348167 | Rat | RGD:1624291 |
| Molecular Function | GO:0042802 | identical protein binding | Cad | IDA | Rat | RGD:2303534|PMID:26565 | |
| Molecular Function | GO:0016874 | ligase activity | pyr1-3 | ISS | InterPro:IPR005481 | Dicty | dictyBase_REF:1660|PMID:2917570 |
| Molecular Function | GO:0046872 | metal ion binding | carB | IDA | Ecoli | PMID:229896 | |
| Molecular Function | GO:0000166 | nucleotide binding | carB | IMP | Ecoli | PMID:10843852 | |
| Molecular Function | GO:0005543 | phospholipid binding | Cps1 | IDA | Rat | RGD:1582379|PMID:3680220 | |
| Molecular Function | GO:0005515 | protein binding | P31327 | IPI | UniProtKB:P04049 | Human | PMID:12620389 |
| Molecular Function | GO:0005515 | protein binding | P31327 | IPI | UniProtKB:P10398 | Human | PMID:12620389 |
| Molecular Function | GO:0005515 | protein binding | Cps1 | ISO | RGD:70833 | Rat | RGD:1624291 |
| Molecular Function | GO:0005515 | protein binding | carB | IPI | UniProtKB:P0A6F1 | Ecoli | PMID:15690043 |
| Molecular Function | GO:0005515 | protein binding | carB | IDA | Ecoli | PMID:4358555 | |
| Molecular Function | GO:0032403 | protein complex binding | Cps1 | IDA | Rat | RGD:2303511|PMID:17669278 | |
| Molecular Function | GO:0004672 | protein kinase activity | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Molecular Function | GO:0002134 | UTP binding | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Cellular Component | GO:0005951 | carbamoyl-phosphate synthase complex | CPA2 | ISS | Yeast | SGD_REF:S000057270|PMID:6358221 | |
| Cellular Component | GO:0009507 | chloroplast | CARB | IDA | Dicot | TAIR:Publication:501712079|PMID:15028209 | |
| Cellular Component | GO:0009507 | chloroplast | CARB | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0009570 | chloroplast stroma | CARB | IDA | Dicot | TAIR:Publication:501728638|PMID:16207701 | |
| Cellular Component | GO:0009570 | chloroplast stroma | CARB | IDA | Dicot | TAIR:Publication:501727324|PMID:18633119 | |
| Cellular Component | GO:0005737 | cytoplasm | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Cellular Component | GO:0005737 | cytoplasm | Cps1 | IDA | Rat | RGD:2303522|PMID:3527129 | |
| Cellular Component | GO:0005737 | cytoplasm | URA2 | IDA | Yeast | SGD_REF:S000059391|PMID:11015727 | |
| Cellular Component | GO:0005737 | cytoplasm | ura1 | ISS | SGD:S000003666 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005829 | cytosol | P27708 | EXP | Human | PMID:8619816 | |
| Cellular Component | GO:0005829 | cytosol | P27708 | IDA | Human | PMID:15890648 | |
| Cellular Component | GO:0005829 | cytosol | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Cellular Component | GO:0005829 | cytosol | Cad | ISO | RGD:1348167 | Rat | RGD:1624291 |
| Cellular Component | GO:0005829 | cytosol | arg4 | ISS | UniProtKB:P03965 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005829 | cytosol | ura1 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0016021 | integral to membrane | URA2 | IDA | Yeast | SGD_REF:S000069514|PMID:11921093 | |
| Cellular Component | GO:0016020 | membrane | CARB | IDA | Dicot | TAIR:Publication:501721401|PMID:17432890 | |
| Cellular Component | GO:0005743 | mitochondrial inner membrane | Cps1 | IDA | Rat | RGD:1582379|PMID:3680220 | |
| Cellular Component | GO:0042645 | mitochondrial nucleoid | P31327 | IDA | Human | PMID:18063578 | |
| Cellular Component | GO:0042645 | mitochondrial nucleoid | Cps1 | ISO | RGD:70833 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | Cps1 | IDA | Mouse | MGI:MGI:3831659|PMID:6200105 | |
| Cellular Component | GO:0005739 | mitochondrion | Cps1 | IDA | Rat | RGD:14561759 | |
| Cellular Component | GO:0005739 | mitochondrion | Cps1 | ISO | RGD:10389 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | URA2 | IDA | Yeast | SGD_REF:S000075100|PMID:14576278 | |
| Cellular Component | GO:0005739 | mitochondrion | URA2 | IDA | Yeast | SGD_REF:S000117178|PMID:16823961 | |
| Cellular Component | GO:0005739 | mitochondrion | ura1 | ISS | SGD:S000003666 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005739 | mitochondrion | arg4 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0016363 | nuclear matrix | P27708 | IDA | Human | PMID:9525610 | |
| Cellular Component | GO:0016363 | nuclear matrix | Cad | ISO | RGD:1348167 | Rat | RGD:1624291 |
| Cellular Component | GO:0005634 | nucleus | P27708 | IDA | Human | PMID:15890648 | |
| Cellular Component | GO:0005634 | nucleus | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Cellular Component | GO:0005634 | nucleus | Cad | ISO | RGD:1348167 | Rat | RGD:1624291 |
| Cellular Component | GO:0043234 | protein complex | Cps1 | IDA | Rat | RGD:2303511|PMID:17669278 | |
| Cellular Component | GO:0043234 | protein complex | Cad | IDA | Rat | RGD:2303534|PMID:26565 | |
| Cellular Component | GO:0005625 | soluble fraction | Cad | IDA | Rat | RGD:2303606|PMID:12803 | |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | cad | IMP | ZFIN:ZDB-GENO-010731-7 | Zfish | ZFIN:ZDB-PUB-050607-20|PMID:15937129 |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | pyr1-3 | ISS | UniProt:P27708 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | URA2 | IMP | Yeast | SGD_REF:S000124960|PMID:4550660 | |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | ura1 | IGI | SGD:S000003666 | Pombe | PMID:17072883 |
| Biological Process | GO:0008652 | amino acid biosynthetic process | carB | IMP | Ecoli | PMID:1737023 | |
| Biological Process | GO:0055081 | anion homeostasis | Cps1 | IDA | Rat | RGD:2300098|PMID:4062872 | |
| Biological Process | GO:0006526 | arginine biosynthetic process | arg4 | ISS | UniProtKB:P03965 | Pombe | PMID:17072883 |
| Biological Process | GO:0006531 | aspartate metabolic process | ura1 | IC | GO:0004070 | Pombe | PMID:17072883 |
| Biological Process | GO:0017144 | drug metabolic process | Cad | IMP | Rat | RGD:2303526|PMID:18515330 | |
| Biological Process | GO:0009790 | embryonic development | Cad | IEP | Rat | RGD:2303538|PMID:14160653 | |
| Biological Process | GO:0009790 | embryonic development | pyr-1 | IMP | WB:cu8|WB:WBPhenotype:0001020 | Worm | WB:WBPaper00031153|PMID:16828468 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | pyr-1 | IMP | WB:WBRNAi00008581|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0007565 | female pregnancy | Cad | IEP | Rat | RGD:2303605|PMID:6020217 | |
| Biological Process | GO:0001703 | gastrulation with mouth forming first | pyr-1 | IMP | WB:cu8|WB:WBPhenotype:0001020 | Worm | WB:WBPaper00031153|PMID:16828468 |
| Biological Process | GO:0006538 | glutamate catabolic process | arg4 | IC | GO:0004088 | Pombe | PMID:17072883 |
| Biological Process | GO:0006541 | glutamine metabolic process | pyr1-3 | IDA | Dicty | dictyBase_REF:6651|PMID:2568162 | |
| Biological Process | GO:0006541 | glutamine metabolic process | URA2 | IDA | Yeast | SGD_REF:S000065623|PMID:5651325 | |
| Biological Process | GO:0006541 | glutamine metabolic process | ura1 | ISS | SGD:S000003666 | Pombe | PMID:17072883 |
| Biological Process | GO:0005980 | glycogen catabolic process | P31327 | IMP | Human | PMID:9711878 | |
| Biological Process | GO:0005980 | glycogen catabolic process | Cps1 | ISO | RGD:70833 | Rat | RGD:1624291 |
| Biological Process | GO:0040011 | locomotion | pyr-1 | IMP | WB:WBRNAi00008581|WB:WBPhenotype:0000643 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0018996 | molting cycle, collagen and cuticulin-based cuticle | pyr-1 | IMP | WB:WBRNAi00008581|WB:WBPhenotype:0000638 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0018996 | molting cycle, collagen and cuticulin-based cuticle | pyr-1 | IMP | WB:WBRNAi00066888|WB:WBPhenotype:0000638 | Worm | WB:WBPaper00026763|PMID:16122351 |
| Biological Process | GO:0002119 | nematode larval development | pyr-1 | IMP | WB:WBRNAi00008581|WB:WBPhenotype:0000054 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0046209 | nitric oxide metabolic process | P31327 | IMP | Human | PMID:14718356 | |
| Biological Process | GO:0046209 | nitric oxide metabolic process | Cps1 | ISO | RGD:70833 | Rat | RGD:1624291 |
| Biological Process | GO:0031100 | organ regeneration | Cad | IEP | Rat | RGD:2303539|PMID:13671431 | |
| Biological Process | GO:0018107 | peptidyl-threonine phosphorylation | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Biological Process | GO:0040010 | positive regulation of growth rate | pyr-1 | IMP | WB:WBRNAi00008581|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0040010 | positive regulation of growth rate | pyr-1 | IMP | WB:cu8|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00031153|PMID:16828468 |
| Biological Process | GO:0045909 | positive regulation of vasodilation | P31327 | IMP | Human | PMID:14718356 | |
| Biological Process | GO:0045909 | positive regulation of vasodilation | Cps1 | ISO | RGD:70833 | Rat | RGD:1624291 |
| Biological Process | GO:0046777 | protein amino acid autophosphorylation | Cad | ISO | UniProtKB:P08955 | Mouse | MGI:MGI:2154458 |
| Biological Process | GO:0019856 | pyrimidine base biosynthetic process | URA2 | IDA | Yeast | SGD_REF:S000054034|PMID:181668 | |
| Biological Process | GO:0019856 | pyrimidine base biosynthetic process | carB | IMP | Ecoli | PMID:1737023 | |
| Biological Process | GO:0014075 | response to amine stimulus | Cad | IEP | Rat | RGD:2303531|PMID:2864015 | |
| Biological Process | GO:0014075 | response to amine stimulus | Cps1 | IEP | Rat | RGD:2303531|PMID:2864015 | |
| Biological Process | GO:0031000 | response to caffeine | Cad | IEP | Rat | RGD:2303533|PMID:6149265 | |
| Biological Process | GO:0051591 | response to cAMP | Cps1 | IEP | Rat | RGD:2303523|PMID:6092398 | |
| Biological Process | GO:0051591 | response to cAMP | Cps1 | IEP | Rat | RGD:2303516|PMID:17397987 | |
| Biological Process | GO:0042493 | response to drug | Cps1 | IEP | Rat | RGD:2303405|PMID:19135993 | |
| Biological Process | GO:0033762 | response to glucagon stimulus | Cps1 | IEP | Rat | RGD:2303521|PMID:3754512 | |
| Biological Process | GO:0051384 | response to glucocorticoid stimulus | Cps1 | IEP | Rat | RGD:2303518|PMID:8985169 | |
| Biological Process | GO:0051384 | response to glucocorticoid stimulus | Cps1 | IEP | Rat | RGD:2303516|PMID:17397987 | |
| Biological Process | GO:0032496 | response to lipopolysaccharide | P31327 | IDA | Human | PMID:15897806 | |
| Biological Process | GO:0032496 | response to lipopolysaccharide | Cps1 | IEP | Rat | RGD:2303515|PMID:17539997 | |
| Biological Process | GO:0032496 | response to lipopolysaccharide | Cps1 | ISO | RGD:70833 | Rat | RGD:1624291 |
| Biological Process | GO:0034201 | response to oleate | Cps1 | IEP | Rat | RGD:2303518|PMID:8985169 | |
| Biological Process | GO:0042594 | response to starvation | Cps1 | IEP | Rat | RGD:2303520|PMID:1979948 | |
| Biological Process | GO:0033574 | response to testosterone stimulus | Cad | IEP | Rat | RGD:2303540|PMID:6030068 | |
| Biological Process | GO:0019433 | triglyceride catabolic process | P31327 | IMP | Human | PMID:9711878 | |
| Biological Process | GO:0019433 | triglyceride catabolic process | Cps1 | ISO | RGD:70833 | Rat | RGD:1624291 |
| Biological Process | GO:0000050 | urea cycle | Cps1 | IC | GO:0004087 | Mouse | MGI:MGI:1328875|PMID:9862865 |
| Biological Process | GO:0000050 | urea cycle | Cps1 | IDA | Rat | RGD:2300098|PMID:4062872 | |
| Biological Process | GO:0000050 | urea cycle | Cps1 | IDA | Rat | RGD:1582379|PMID:3680220 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |