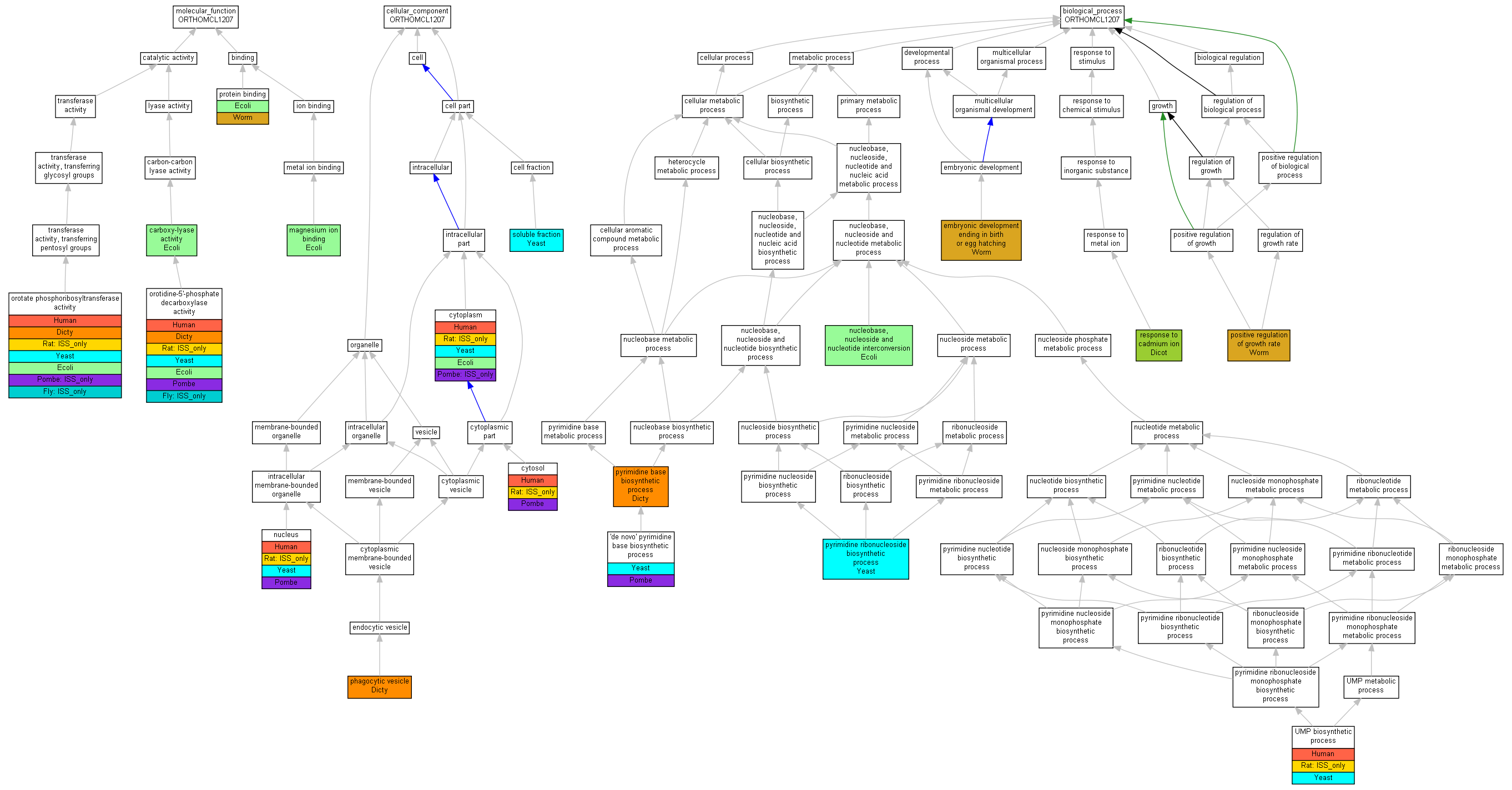
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | (Rat) | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016831 | carboxy-lyase activity | pyrF | IDA | Ecoli | PMID:6355062 | |
| Molecular Function | GO:0000287 | magnesium ion binding | pyrE | IDA | Ecoli | PMID:7050303 | |
| Molecular Function | GO:0000287 | magnesium ion binding | pyrE | IDA | Ecoli | PMID:8620002 | |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | P11172 | IDA | Human | PMID:6893554 | |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | P11172 | EXP | Human | PMID:9042911 | |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | P11172 | IDA | Human | PMID:11730338 | |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | Umps | ISO | RGD:1323612 | Rat | RGD:1624291 |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | r-l | ISS | NCBI_gi:4507835 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | pyr56 | ISS | Dicty | dictyBase_REF:2869|PMID:2835631 | |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | pyr56 | IGI | SGD:S000004574 | Dicty | dictyBase_REF:2869|PMID:2835631 |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | pyr56 | IGI | SGD:S000004574 | Dicty | dictyBase_REF:5709|PMID:8500761 |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | URA10 | IMP | Yeast | SGD_REF:S000048197|PMID:2182197 | |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | URA10 | ISA | SGD:S000004574 | Yeast | SGD_REF:S000048197|PMID:2182197 |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | URA10 | IGI | SGD:S000004574 | Yeast | SGD_REF:S000048197|PMID:2182197 |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | URA5 | IDA | Yeast | SGD_REF:S000126381|PMID:9882434 | |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | URA5 | IMP | Yeast | SGD_REF:S000124960|PMID:4550660 | |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | ura5 | ISS | SGD:S000004574 | Pombe | PMID:17072883 |
| Molecular Function | GO:0004588 | orotate phosphoribosyltransferase activity | pyrE | IDA | Ecoli | PMID:9222315 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | P11172 | EXP | Human | PMID:9042911 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | P11172 | IDA | Human | PMID:11730338 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | P11172 | IDA | Human | PMID:6893554 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | Umps | ISO | RGD:1323612 | Rat | RGD:1624291 |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | r-l | ISS | NCBI_gi:4507835 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | pyr56 | IGI | SGD:S000000747 | Dicty | dictyBase_REF:2869|PMID:2835631 |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | pyr56 | ISS | Dicty | dictyBase_REF:2869|PMID:2835631 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | pyr56 | IGI | SGD:S000000747 | Dicty | dictyBase_REF:5709|PMID:8500761 |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | pyr56 | IMP | Dicty | dictyBase_REF:3018|PMID:2017144 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | URA3 | IMP | Yeast | SGD_REF:S000124960|PMID:4550660 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | URA3 | IDA | Yeast | SGD_REF:S000052912|PMID:2061334 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | URA3 | IMP | Yeast | SGD_REF:S000065623|PMID:5651325 | |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | ura4 | IGI | UniProtKB:PYRF_ECOLI | Pombe | PMID:2834100 |
| Molecular Function | GO:0004590 | orotidine-5'-phosphate decarboxylase activity | pyrF | IDA | Ecoli | PMID:6355062 | |
| Molecular Function | GO:0005515 | protein binding | umps-1 | IPI | UniProtKB:O17218 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | umps-1 | IPI | UniProtKB:O18180 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | umps-1 | IPI | UniProtKB:O45784 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | umps-1 | IPI | UniProtKB:P48154 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | umps-1 | IPI | UniProtKB:Q18809 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | umps-1 | IPI | UniProtKB:Q18869 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | umps-1 | IPI | UniProtKB:Q22003 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | umps-1 | IPI | UniProtKB:Q9XWW1 | Worm | PMID:14704431 |
| Molecular Function | GO:0005515 | protein binding | pyrE | IPI | UniProtKB:P0A6B7 | Ecoli | PMID:16606699 |
| Molecular Function | GO:0005515 | protein binding | pyrF | IPI | UniProtKB:P0A7I7 | Ecoli | PMID:16606699 |
| Cellular Component | GO:0005737 | cytoplasm | P11172 | IDA | Human | PMID:15890648 | |
| Cellular Component | GO:0005737 | cytoplasm | Umps | ISO | RGD:1323612 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | URA10 | IDA | Yeast | SGD_REF:S000074185|PMID:14562095 | |
| Cellular Component | GO:0005737 | cytoplasm | URA5 | IDA | Yeast | SGD_REF:S000069459|PMID:11914276 | |
| Cellular Component | GO:0005737 | cytoplasm | URA5 | IDA | Yeast | SGD_REF:S000074185|PMID:14562095 | |
| Cellular Component | GO:0005737 | cytoplasm | ura5 | ISS | SGD:S000004574 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005737 | cytoplasm | pyrE | IDA | Ecoli | PMID:9222315 | |
| Cellular Component | GO:0005737 | cytoplasm | pyrF | IDA | Ecoli | PMID:6355062 | |
| Cellular Component | GO:0005829 | cytosol | P11172 | EXP | Human | PMID:9042911 | |
| Cellular Component | GO:0005829 | cytosol | Umps | ISO | RGD:1323612 | Rat | RGD:1624291 |
| Cellular Component | GO:0005829 | cytosol | ura4 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005829 | cytosol | ura5 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005634 | nucleus | P11172 | IDA | Human | PMID:15890648 | |
| Cellular Component | GO:0005634 | nucleus | Umps | ISO | RGD:1323612 | Rat | RGD:1624291 |
| Cellular Component | GO:0005634 | nucleus | URA5 | IDA | Yeast | SGD_REF:S000074185|PMID:14562095 | |
| Cellular Component | GO:0005634 | nucleus | ura5 | ISS | SGD:S000004574 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005634 | nucleus | ura4 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005634 | nucleus | ura5 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0045335 | phagocytic vesicle | pyr56 | IDA | Dicty | dictyBase_REF:11413|PMID:16926386 | |
| Cellular Component | GO:0005625 | soluble fraction | URA3 | IDA | Yeast | SGD_REF:S000065638|PMID:374408 | |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | URA10 | IGI | SGD:S000004574 | Yeast | SGD_REF:S000048197|PMID:2182197 |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | URA3 | IMP | Yeast | SGD_REF:S000065623|PMID:5651325 | |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | URA3 | IMP | Yeast | SGD_REF:S000124960|PMID:4550660 | |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | URA5 | IMP | Yeast | SGD_REF:S000124960|PMID:4550660 | |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | ura4 | IGI | UniProtKB:PYRF_ECOLI | Pombe | PMID:2834100 |
| Biological Process | GO:0006207 | 'de novo' pyrimidine base biosynthetic process | ura5 | ISS | SGD:S000004574 | Pombe | PMID:17072883 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | R12E2.11 | IMP | WB:WBRNAi00026105|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0015949 | nucleobase, nucleoside and nucleotide interconversion | pyrF | IDA | Ecoli | PMID:6355062 | |
| Biological Process | GO:0040010 | positive regulation of growth rate | umps-1 | IMP | WB:WBRNAi00009137|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0040010 | positive regulation of growth rate | umps-1 | IMP | WB:WBRNAi00026225|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0019856 | pyrimidine base biosynthetic process | pyr56 | IGI | SGD:S000000747 | Dicty | dictyBase_REF:2869|PMID:2835631 |
| Biological Process | GO:0046132 | pyrimidine ribonucleoside biosynthetic process | URA10 | IMP | Yeast | SGD_REF:S000048197|PMID:2182197 | |
| Biological Process | GO:0046132 | pyrimidine ribonucleoside biosynthetic process | URA10 | IGI | SGD:S000004574 | Yeast | SGD_REF:S000048197|PMID:2182197 |
| Biological Process | GO:0046132 | pyrimidine ribonucleoside biosynthetic process | URA5 | IMP | Yeast | SGD_REF:S000124960|PMID:4550660 | |
| Biological Process | GO:0046132 | pyrimidine ribonucleoside biosynthetic process | URA5 | IDA | Yeast | SGD_REF:S000126381|PMID:9882434 | |
| Biological Process | GO:0046686 | response to cadmium ion | AT3G54470.1 | IEP | Dicot | TAIR:Publication:501718481|PMID:16502469 | |
| Biological Process | GO:0006222 | UMP biosynthetic process | P11172 | IDA | Human | PMID:6893554 | |
| Biological Process | GO:0006222 | UMP biosynthetic process | P11172 | IDA | Human | PMID:11730338 | |
| Biological Process | GO:0006222 | UMP biosynthetic process | Umps | ISO | RGD:1323612 | Rat | RGD:1624291 |
| Biological Process | GO:0006222 | UMP biosynthetic process | URA3 | IDA | Yeast | SGD_REF:S000052912|PMID:2061334 | |
| Biological Process | GO:0006222 | UMP biosynthetic process | URA3 | IMP | Yeast | SGD_REF:S000124960|PMID:4550660 | |
| Biological Process | GO:0006222 | UMP biosynthetic process | URA3 | IMP | Yeast | SGD_REF:S000065623|PMID:5651325 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |