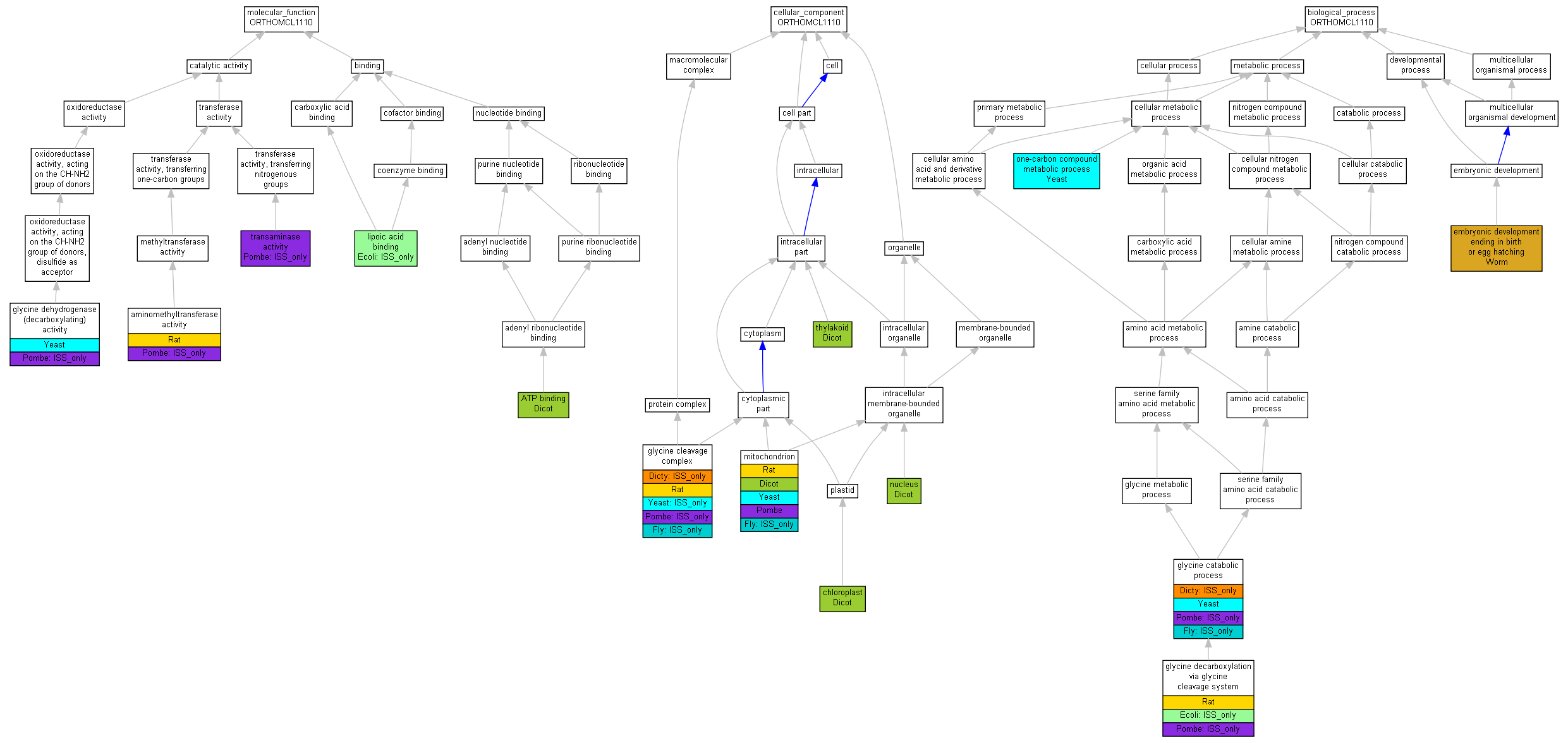
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | (Ecoli) |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0032991 | macromolecular complex | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | (Yeast) | (Pombe) | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | (Ecoli) |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | (Ecoli) |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004047 | aminomethyltransferase activity | Gcsh | IMP | Rat | RGD:1598699|PMID:6402507 | |
| Molecular Function | GO:0004047 | aminomethyltransferase activity | SPBP19A11.01 | ISS | UniProtKB:P39726 | Pombe | PMID:17072883 |
| Molecular Function | GO:0005524 | ATP binding | AT2G35120.1 | IDA | Dicot | TAIR:Publication:501720283|PMID:17137349 | |
| Molecular Function | GO:0004375 | glycine dehydrogenase (decarboxylating) activity | GCV3 | IMP | Yeast | SGD_REF:S000042313|PMID:9020168 | |
| Molecular Function | GO:0004375 | glycine dehydrogenase (decarboxylating) activity | GCV3 | ISA | UniProtKB:P16048 | Yeast | SGD_REF:S000042313|PMID:9020168 |
| Molecular Function | GO:0004375 | glycine dehydrogenase (decarboxylating) activity | SPBP19A11.01 | ISS | UniProtKB:P39726 | Pombe | PMID:17072883 |
| Molecular Function | GO:0031405 | lipoic acid binding | gcvH | ISS | UniProtKB:P11183 | Ecoli | PMID:1802033 |
| Molecular Function | GO:0008483 | transaminase activity | SPBP19A11.01 | ISS | UniProtKB:P39726 | Pombe | PMID:17072883 |
| Cellular Component | GO:0009507 | chloroplast | AT1G32470.1 | IDA | Dicot | TAIR:Publication:501712079|PMID:15028209 | |
| Cellular Component | GO:0009507 | chloroplast | GDCH | IDA | Dicot | TAIR:Publication:501712079|PMID:15028209 | |
| Cellular Component | GO:0005960 | glycine cleavage complex | Gcsh | IC | GO:0019464 | Rat | RGD:1598699|PMID:6402507 |
| Cellular Component | GO:0005960 | glycine cleavage complex | ppl | ISS | NCBI_gi:4758424 | Fly | FB:FBrf0105495 |
| Cellular Component | GO:0005960 | glycine cleavage complex | ppl | ISS | Fly | FB:FBrf0122457 | |
| Cellular Component | GO:0005960 | glycine cleavage complex | ppl | ISS | UniProtKB:P23434 | Fly | FB:FBrf0159903 |
| Cellular Component | GO:0005960 | glycine cleavage complex | gcvH1 | ISS | InterPro:IPR002930 | Dicty | dictyBase_REF:10155 |
| Cellular Component | GO:0005960 | glycine cleavage complex | GCV3 | ISA | UniProtKB:P16048 | Yeast | SGD_REF:S000042313|PMID:9020168 |
| Cellular Component | GO:0005960 | glycine cleavage complex | SPBP19A11.01 | ISS | Pfam:PF01597 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005739 | mitochondrion | Gcsh | IDA | Rat | RGD:1598699|PMID:6402507 | |
| Cellular Component | GO:0005739 | mitochondrion | ppl | ISS | Fly | FB:FBrf0131806 | |
| Cellular Component | GO:0005739 | mitochondrion | ppl | ISS | UniProtKB:P23434 | Fly | FB:FBrf0159903 |
| Cellular Component | GO:0005739 | mitochondrion | AT2G35120.1 | IDA | Dicot | TAIR:Publication:501720283|PMID:17137349 | |
| Cellular Component | GO:0005739 | mitochondrion | AT1G32470.1 | IDA | Dicot | TAIR:Publication:501724621|PMID:18385124 | |
| Cellular Component | GO:0005739 | mitochondrion | AT2G35120.1 | IDA | Dicot | TAIR:Publication:501724621|PMID:18385124 | |
| Cellular Component | GO:0005739 | mitochondrion | GCV3 | IDA | Yeast | SGD_REF:S000075100|PMID:14576278 | |
| Cellular Component | GO:0005739 | mitochondrion | GCV3 | IDA | Yeast | SGD_REF:S000117178|PMID:16823961 | |
| Cellular Component | GO:0005739 | mitochondrion | SPBP19A11.01 | ISS | UniProtKB:P39726 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005739 | mitochondrion | SPBP19A11.01 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005634 | nucleus | GDCH | IDA | Dicot | TAIR:Publication:501711432|PMID:14617066 | |
| Cellular Component | GO:0009579 | thylakoid | GDCH | IDA | Dicot | TAIR:Publication:501681963|PMID:11826309 | |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | F52A8.5 | IMP | WB:WBRNAi00001643|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00004651|PMID:11231151 |
| Biological Process | GO:0006546 | glycine catabolic process | ppl | ISS | Fly | FB:FBrf0122457 | |
| Biological Process | GO:0006546 | glycine catabolic process | ppl | ISS | UniProtKB:P23434 | Fly | FB:FBrf0159903 |
| Biological Process | GO:0006546 | glycine catabolic process | gcvH1 | ISS | InterPro:IPR002930 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006546 | glycine catabolic process | GCV3 | IMP | Yeast | SGD_REF:S000042313|PMID:9020168 | |
| Biological Process | GO:0006546 | glycine catabolic process | SPBP19A11.01 | ISS | Pfam:PF01597 | Pombe | PMID:17072883 |
| Biological Process | GO:0019464 | glycine decarboxylation via glycine cleavage system | Gcsh | IMP | Rat | RGD:1598699|PMID:6402507 | |
| Biological Process | GO:0019464 | glycine decarboxylation via glycine cleavage system | SPBP19A11.01 | ISS | UniProtKB:P39726 | Pombe | PMID:17072883 |
| Biological Process | GO:0019464 | glycine decarboxylation via glycine cleavage system | gcvH | ISS | UniProtKB:P11183 | Ecoli | PMID:1802033 |
| Biological Process | GO:0006730 | one-carbon compound metabolic process | GCV3 | IGI | SGD:S000004048 | Yeast | SGD_REF:S000056431|PMID:10871621 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |