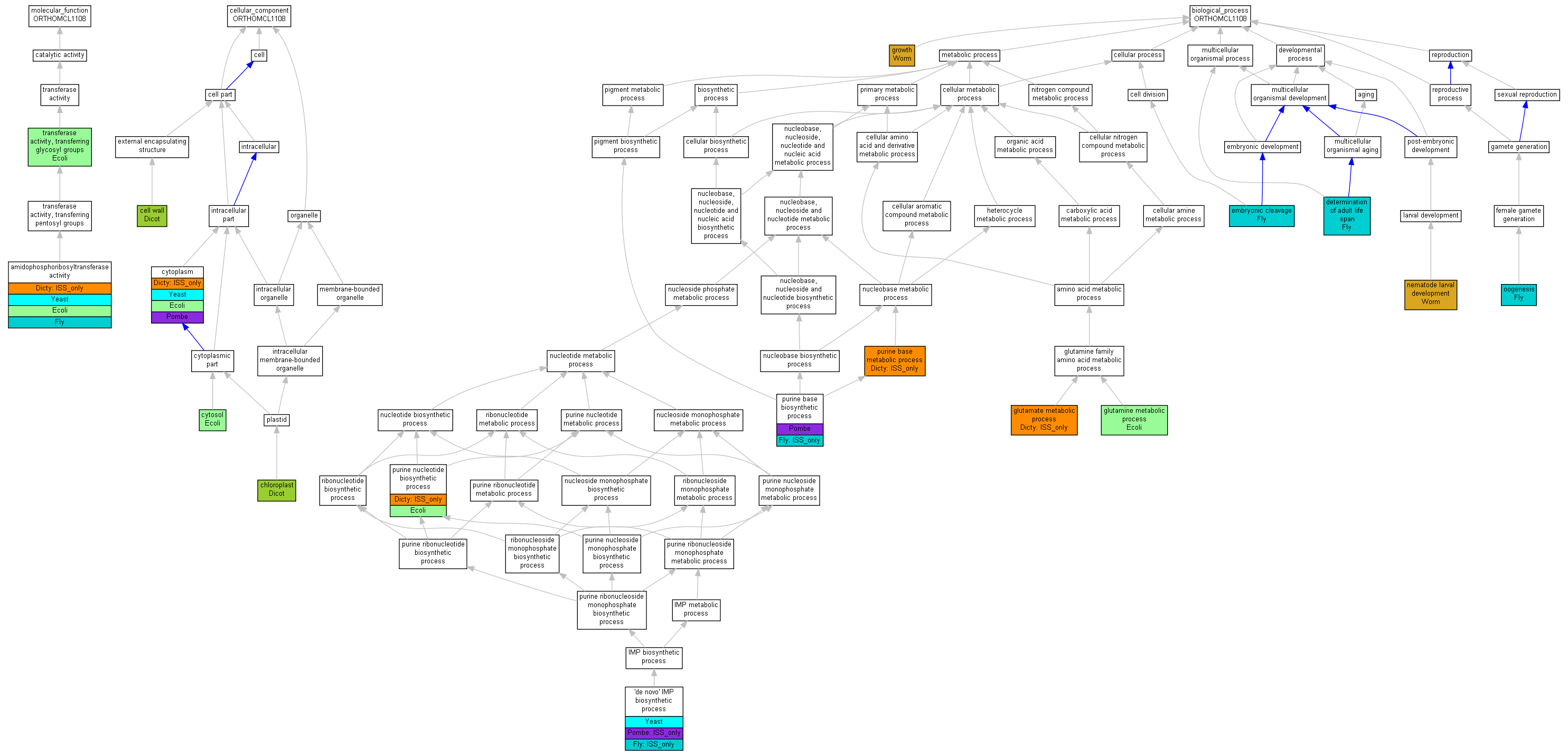
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0000003 | reproduction | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0022414 | reproductive process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | Prat | IMP | Fly | FB:FBrf0068452|PMID:8150282 | |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | Prat | ISS | Fly | FB:FBrf0068452|PMID:8150282 | |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | Prat | ISS | UniProtKB:P28173 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | Prat2 | ISS | UniProtKB:P28173 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | purF | ISS | SGD:S000004915 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | ADE4 | IMP | Yeast | SGD_REF:S000068861|PMID:11689683 | |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | ADE4 | IDA | Yeast | SGD_REF:S000068861|PMID:11689683 | |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | ADE4 | IMP | Yeast | SGD_REF:S000045527|PMID:6376509 | |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | purF | IDA | Ecoli | PMID:372191 | |
| Molecular Function | GO:0004044 | amidophosphoribosyltransferase activity | purF | IDA | Ecoli | PMID:7037784 | |
| Molecular Function | GO:0016757 | transferase activity, transferring glycosyl groups | purF | IDA | Ecoli | PMID:7037784 | |
| Cellular Component | GO:0005618 | cell wall | ATASE1 | IDA | Dicot | TAIR:Publication:501718077|PMID:16287169 | |
| Cellular Component | GO:0009507 | chloroplast | ATASE2 | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0005737 | cytoplasm | purF | ISS | SGD:S000004915 | Dicty | dictyBase_REF:10155 |
| Cellular Component | GO:0005737 | cytoplasm | ADE4 | IDA | Yeast | SGD_REF:S000074185|PMID:14562095 | |
| Cellular Component | GO:0005737 | cytoplasm | ade4 | ISS | UniProtKB:P04046 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005737 | cytoplasm | ade4 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005737 | cytoplasm | purF | IDA | Ecoli | PMID:7037784 | |
| Cellular Component | GO:0005829 | cytosol | purF | IDA | Ecoli | PMID:16858726 | |
| Biological Process | GO:0006189 | 'de novo' IMP biosynthetic process | Prat2 | ISS | Fly | FB:FBrf0105495 | |
| Biological Process | GO:0006189 | 'de novo' IMP biosynthetic process | ADE4 | IMP | Yeast | SGD_REF:S000068861|PMID:11689683 | |
| Biological Process | GO:0006189 | 'de novo' IMP biosynthetic process | ade4 | ISS | SGD:S000004915 | Pombe | PMID:17072883 |
| Biological Process | GO:0008340 | determination of adult life span | Prat | IMP | Fly | FB:FBrf0183154|PMID:15611171 | |
| Biological Process | GO:0040016 | embryonic cleavage | Prat | IMP | Fly | FB:FBrf0183154|PMID:15611171 | |
| Biological Process | GO:0006536 | glutamate metabolic process | purF | ISS | KEGG_PATHWAY:MAP00251 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006541 | glutamine metabolic process | purF | IDA | Ecoli | PMID:7037784 | |
| Biological Process | GO:0040007 | growth | T04A8.5 | IMP | WB:WBRNAi00002700|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00004403|PMID:11099034 |
| Biological Process | GO:0002119 | nematode larval development | T04A8.5 | IMP | WB:WBRNAi00002700|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00004403|PMID:11099034 |
| Biological Process | GO:0048477 | oogenesis | Prat | IMP | Fly | FB:FBrf0183154|PMID:15611171 | |
| Biological Process | GO:0009113 | purine base biosynthetic process | Prat | ISS | Fly | FB:FBrf0068452|PMID:8150282 | |
| Biological Process | GO:0009113 | purine base biosynthetic process | ade4 | IC | GO:0004044 | Pombe | PMID:17072883 |
| Biological Process | GO:0006144 | purine base metabolic process | purF | ISS | SGD:S000004915 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006164 | purine nucleotide biosynthetic process | purF | ISS | SGD:S000004915 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006164 | purine nucleotide biosynthetic process | purF | IDA | Ecoli | PMID:7037784 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |