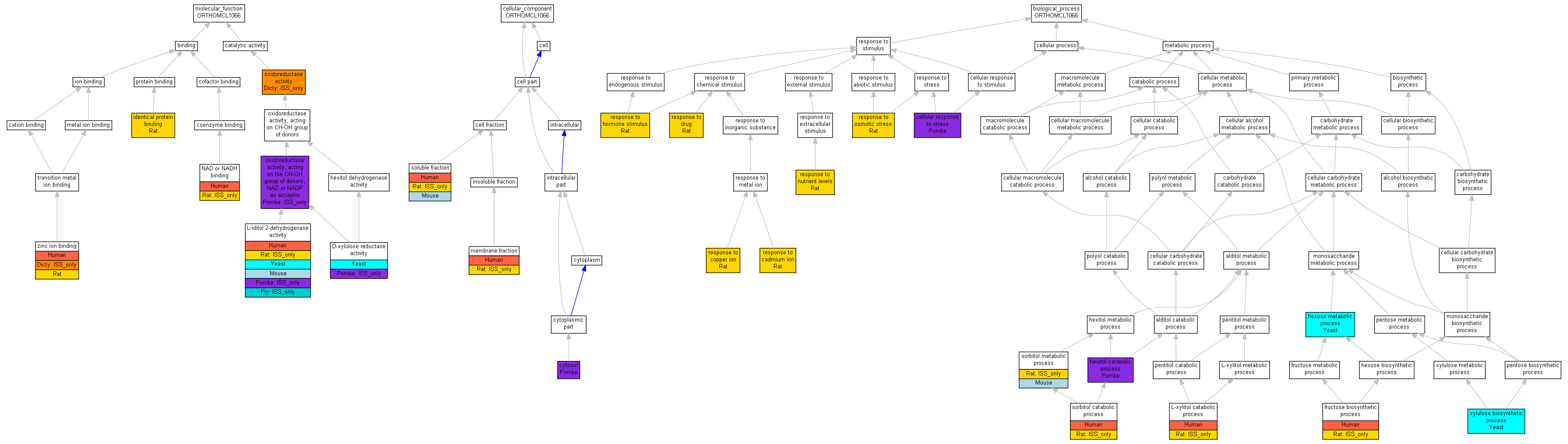
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | (Rat) | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0046526 | D-xylulose reductase activity | XYL2 | IDA | Yeast | SGD_REF:S000056333|PMID:10486580 | |
| Molecular Function | GO:0046526 | D-xylulose reductase activity | tms1 | ISS | SGD:S000004060 | Pombe | PMID:17072883 |
| Molecular Function | GO:0042802 | identical protein binding | Sord | IDA | Rat | RGD:1601367|PMID:11306057 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Q00796 | IDA | Human | PMID:3365415 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Q00796 | IDA | Human | PMID:6870831 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Q00796 | IDA | Human | PMID:8487505 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Q00796 | ISS | UniProtKB:P07846 | Human | GO_REF:0000024 |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Sord | IDA | Mouse | MGI:MGI:48978|PMID:1914521 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Sord | IDA | Mouse | MGI:MGI:55526|PMID:7171128 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Sord | IDA | Mouse | MGI:MGI:55679|PMID:6626150 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Sord | IDA | Mouse | MGI:MGI:60299|PMID:722274 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Sord | ISO | RGD:11332 | Rat | RGD:1624291 |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Sord | ISO | RGD:735508 | Rat | RGD:1624291 |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | Sodh-2 | ISS | UniProtKB:Q64442 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | SOR1 | ISS | UniProtKB:P07846 | Yeast | SGD_REF:S000057608|PMID:8125328 |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | SOR1 | IDA | Yeast | SGD_REF:S000057608|PMID:8125328 | |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | SOR2 | ISS | SGD:S000003920 | Yeast | SGD_REF:S000056335|PMID:10938079 |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity | tms1 | ISS | SGD:S000003920 | Pombe | PMID:17072883 |
| Molecular Function | GO:0051287 | NAD or NADH binding | Q00796 | IDA | Human | PMID:12962626 | |
| Molecular Function | GO:0051287 | NAD or NADH binding | Sord | ISO | RGD:735508 | Rat | RGD:1624291 |
| Molecular Function | GO:0016491 | oxidoreductase activity | DDB_G0280997 | ISS | InterPro:IPR002085 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | tms1 | ISS | SGD:S00000240 | Pombe | PMID:17072883 |
| Molecular Function | GO:0008270 | zinc ion binding | Q00796 | ISS | UniProtKB:P07846 | Human | GO_REF:0000024 |
| Molecular Function | GO:0008270 | zinc ion binding | Q00796 | IDA | Human | PMID:12962626 | |
| Molecular Function | GO:0008270 | zinc ion binding | Sord | IDA | Rat | RGD:1601367|PMID:11306057 | |
| Molecular Function | GO:0008270 | zinc ion binding | Sord | ISO | RGD:735508 | Rat | RGD:1624291 |
| Molecular Function | GO:0008270 | zinc ion binding | DDB_G0280997 | ISS | InterPro:IPR002085 | Dicty | dictyBase_REF:10155 |
| Cellular Component | GO:0005829 | cytosol | tms1 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005624 | membrane fraction | Q00796 | IDA | Human | PMID:8487505 | |
| Cellular Component | GO:0005624 | membrane fraction | Sord | ISO | RGD:735508 | Rat | RGD:1624291 |
| Cellular Component | GO:0005625 | soluble fraction | Q00796 | IDA | Human | PMID:8487505 | |
| Cellular Component | GO:0005625 | soluble fraction | Q00796 | ISS | UniProtKB:Q64442 | Human | GO_REF:0000024 |
| Cellular Component | GO:0005625 | soluble fraction | Sord | IDA | Mouse | MGI:MGI:55679|PMID:6626150 | |
| Cellular Component | GO:0005625 | soluble fraction | Sord | ISO | RGD:735508 | Rat | RGD:1624291 |
| Cellular Component | GO:0005625 | soluble fraction | Sord | ISO | RGD:11332 | Rat | RGD:1624291 |
| Biological Process | GO:0033554 | cellular response to stress | tms1 | IEP | GOC:pombekw2GO | Pombe | PMID:12529438 |
| Biological Process | GO:0046370 | fructose biosynthetic process | Q00796 | IDA | Human | PMID:8487505 | |
| Biological Process | GO:0046370 | fructose biosynthetic process | Q00796 | IDA | Human | PMID:6870831 | |
| Biological Process | GO:0046370 | fructose biosynthetic process | Q00796 | IDA | Human | PMID:3365415 | |
| Biological Process | GO:0046370 | fructose biosynthetic process | Sord | ISO | RGD:735508 | Rat | RGD:1624291 |
| Biological Process | GO:0019407 | hexitol catabolic process | tms1 | IC | GO:0003939 | Pombe | PMID:17072883 |
| Biological Process | GO:0019318 | hexose metabolic process | SOR1 | IEP | Yeast | SGD_REF:S000057608|PMID:8125328 | |
| Biological Process | GO:0019318 | hexose metabolic process | SOR2 | ISS | SGD:S000003920 | Yeast | SGD_REF:S000056335|PMID:10938079 |
| Biological Process | GO:0051160 | L-xylitol catabolic process | Q00796 | IDA | Human | PMID:8487505 | |
| Biological Process | GO:0051160 | L-xylitol catabolic process | Sord | ISO | RGD:735508 | Rat | RGD:1624291 |
| Biological Process | GO:0046686 | response to cadmium ion | Sord | IDA | Rat | RGD:1601368|PMID:10620479 | |
| Biological Process | GO:0046688 | response to copper ion | Sord | IDA | Rat | RGD:1599065|PMID:11044632 | |
| Biological Process | GO:0042493 | response to drug | Sord | IEP | Rat | RGD:1601360|PMID:15064821 | |
| Biological Process | GO:0009725 | response to hormone stimulus | Sord | IDA | Rat | RGD:1601370|PMID:9838221 | |
| Biological Process | GO:0031667 | response to nutrient levels | Sord | IEP | Rat | RGD:1601361|PMID:16740392 | |
| Biological Process | GO:0006970 | response to osmotic stress | Sord | IEP | Rat | RGD:1601363|PMID:14551351 | |
| Biological Process | GO:0006062 | sorbitol catabolic process | Q00796 | IDA | Human | PMID:8487505 | |
| Biological Process | GO:0006062 | sorbitol catabolic process | Q00796 | IDA | Human | PMID:6870831 | |
| Biological Process | GO:0006062 | sorbitol catabolic process | Q00796 | IDA | Human | PMID:3365415 | |
| Biological Process | GO:0006062 | sorbitol catabolic process | Q00796 | IDA | Human | PMID:12962626 | |
| Biological Process | GO:0006062 | sorbitol catabolic process | Sord | ISO | RGD:735508 | Rat | RGD:1624291 |
| Biological Process | GO:0006060 | sorbitol metabolic process | Sord | IDA | Mouse | MGI:MGI:55679|PMID:6626150 | |
| Biological Process | GO:0006060 | sorbitol metabolic process | Sord | IDA | Mouse | MGI:MGI:60299|PMID:722274 | |
| Biological Process | GO:0006060 | sorbitol metabolic process | Sord | ISO | RGD:11332 | Rat | RGD:1624291 |
| Biological Process | GO:0005999 | xylulose biosynthetic process | XYL2 | IDA | Yeast | SGD_REF:S000056333|PMID:10486580 | |
| Biological Process | GO:0005999 | xylulose biosynthetic process | XYL2 | IEP | Yeast | SGD_REF:S000056333|PMID:10486580 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |