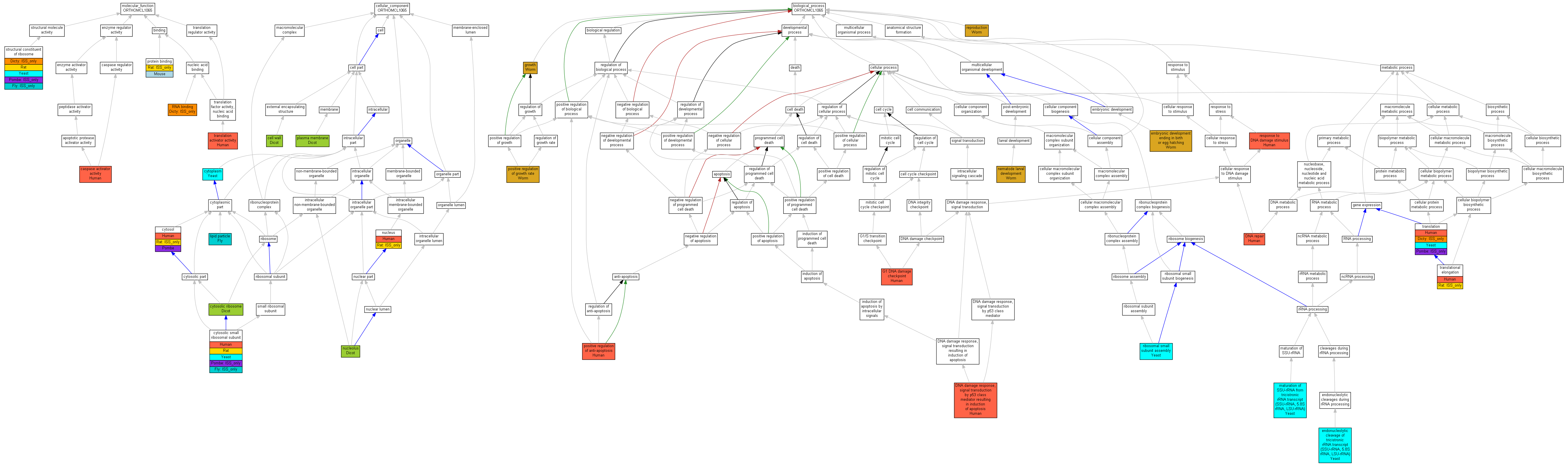
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | X |
| Molecular Function | GO:0030234 | enzyme regulator activity | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Molecular Function | GO:0005198 | structural molecule activity | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Molecular Function | GO:0045182 | translation regulator activity | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0032991 | macromolecular complex | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | X |
| Cellular Component | GO:0031974 | membrane-enclosed lumen | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | X |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0010926 | anatomical structure formation | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0048519 | negative regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0000003 | reproduction | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008656 | caspase activator activity | Q71UM5 | IDA | Human | PMID:17057733 | |
| Molecular Function | GO:0005515 | protein binding | Rps27 | IPI | UniProtKB:Q61937 | Mouse | MGI:MGI:3573985|PMID:15596447 |
| Molecular Function | GO:0005515 | protein binding | Rps27 | ISO | RGD:737001 | Rat | RGD:1624291 |
| Molecular Function | GO:0003723 | RNA binding | rps27 | ISS | UniProt:P42677 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | Rps27 | IDA | Rat | RGD:634035|PMID:10820185 | |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RpS27 | ISS | SGD:S0001063 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | rps27 | ISS | UniProt:P42677 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RPS27A | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Molecular Function | GO:0003735 | structural constituent of ribosome | RPS27B | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Molecular Function | GO:0003735 | structural constituent of ribosome | rps27 | ISS | SGD:S000001639 | Pombe | PMID:17072883 |
| Molecular Function | GO:0008494 | translation activator activity | Q71UM5 | IDA | Human | PMID:18056458 | |
| Cellular Component | GO:0005618 | cell wall | ARS27A | IDA | Dicot | TAIR:Publication:501718077|PMID:16287169 | |
| Cellular Component | GO:0005737 | cytoplasm | RPS27A | IDA | Yeast | SGD_REF:S000069459|PMID:11914276 | |
| Cellular Component | GO:0005829 | cytosol | P42677 | EXP | Human | PMID:12588972 | |
| Cellular Component | GO:0005829 | cytosol | Rps27 | ISO | RGD:732460 | Rat | RGD:1624291 |
| Cellular Component | GO:0005829 | cytosol | rps27 | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | ARS27A | IDA | Dicot | TAIR:Publication:501714926|PMID:15734919 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | AT2G45710.1 | IDA | Dicot | TAIR:Publication:501714975|PMID:15821981 | |
| Cellular Component | GO:0022626 | cytosolic ribosome | AT5G47930.1 | IDA | Dicot | TAIR:Publication:501714975|PMID:15821981 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | P42677 | IDA | Human | PMID:8706699 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | P42677 | IDA | Human | PMID:15883184 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | Rps27 | IDA | Rat | RGD:633964|PMID:8441676 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | Rps27 | ISO | RGD:732460 | Rat | RGD:1624291 |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | Rps27 | IDA | Rat | RGD:2300010|PMID:947902 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RpS27 | ISS | SGD:S0001063 | Fly | FB:FBrf0105495 |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RPS27A | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | RPS27B | IDA | Yeast | SGD_REF:S000116584|PMID:3533916 | |
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | rps27 | ISS | SGD:S000001639 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005811 | lipid particle | RpS27 | IDA | Fly | FB:FBrf0192764|PMID:16979555 | |
| Cellular Component | GO:0005730 | nucleolus | AT2G45710.1 | IDA | Dicot | TAIR:Publication:501714224|PMID:15496452 | |
| Cellular Component | GO:0005634 | nucleus | Q71UM5 | IDA | Human | PMID:18056458 | |
| Cellular Component | GO:0005634 | nucleus | P42677 | IDA | Human | PMID:8407955 | |
| Cellular Component | GO:0005634 | nucleus | Rps27 | ISO | RGD:732460 | Rat | RGD:1624291 |
| Cellular Component | GO:0005886 | plasma membrane | AT2G45710.1 | IDA | Dicot | TAIR:Publication:501720929|PMID:17317660 | |
| Cellular Component | GO:0005886 | plasma membrane | AT5G47930.1 | IDA | Dicot | TAIR:Publication:501720929|PMID:17317660 | |
| Biological Process | GO:0042771 | DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis | Q71UM5 | IDA | Human | PMID:17057733 | |
| Biological Process | GO:0006281 | DNA repair | Q71UM5 | IMP | Human | PMID:18056458 | |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | rps-27 | IMP | WB:WBRNAi00048729|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00025054|PMID:15791247 |
| Biological Process | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | RPS27B | IMP | Yeast | SGD_REF:S000055418|PMID:9271380 | |
| Biological Process | GO:0031571 | G1 DNA damage checkpoint | Q71UM5 | IMP | Human | PMID:18056458 | |
| Biological Process | GO:0040007 | growth | rps-27 | IMP | WB:WBRNAi00025708|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0040007 | growth | rps-27 | IMP | WB:WBRNAi00032965|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00024497|PMID:15489339 |
| Biological Process | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | RPS27A | IGI | SGD:S000001063 | Yeast | SGD_REF:S000087225|PMID:16246728 |
| Biological Process | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | RPS27B | IGI | SGD:S000001639 | Yeast | SGD_REF:S000087225|PMID:16246728 |
| Biological Process | GO:0002119 | nematode larval development | rps-27 | IMP | WB:WBRNAi00025708|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0002119 | nematode larval development | rps-27 | IMP | WB:WBRNAi00032965|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00024497|PMID:15489339 |
| Biological Process | GO:0045768 | positive regulation of anti-apoptosis | Q71UM5 | IMP | Human | PMID:18056458 | |
| Biological Process | GO:0040010 | positive regulation of growth rate | rps-27 | IMP | WB:WBRNAi00008881|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0000003 | reproduction | rps-27 | IMP | WB:WBRNAi00008881|WB:WBPhenotype:0001037 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0006974 | response to DNA damage stimulus | Q71UM5 | IDA | Human | PMID:18056458 | |
| Biological Process | GO:0000028 | ribosomal small subunit assembly | RPS27A | IMP | Yeast | SGD_REF:S000055418|PMID:9271380 | |
| Biological Process | GO:0000028 | ribosomal small subunit assembly | RPS27B | IMP | Yeast | SGD_REF:S000055418|PMID:9271380 | |
| Biological Process | GO:0006412 | translation | Q71UM5 | IDA | Human | PMID:18056458 | |
| Biological Process | GO:0006412 | translation | rps27 | ISS | UniProt:P42677 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006412 | translation | RPS27A | IC | GO:0022627 | Yeast | SGD_REF:S000116584|PMID:3533916 |
| Biological Process | GO:0006412 | translation | RPS27B | IC | GO:0022627 | Yeast | SGD_REF:S000116584|PMID:3533916 |
| Biological Process | GO:0006412 | translation | rps27 | ISS | SGD:S000001639 | Pombe | PMID:17072883 |
| Biological Process | GO:0006414 | translational elongation | P42677 | EXP | Human | PMID:15189156 | |
| Biological Process | GO:0006414 | translational elongation | Rps27 | ISO | RGD:732460 | Rat | RGD:1624291 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |