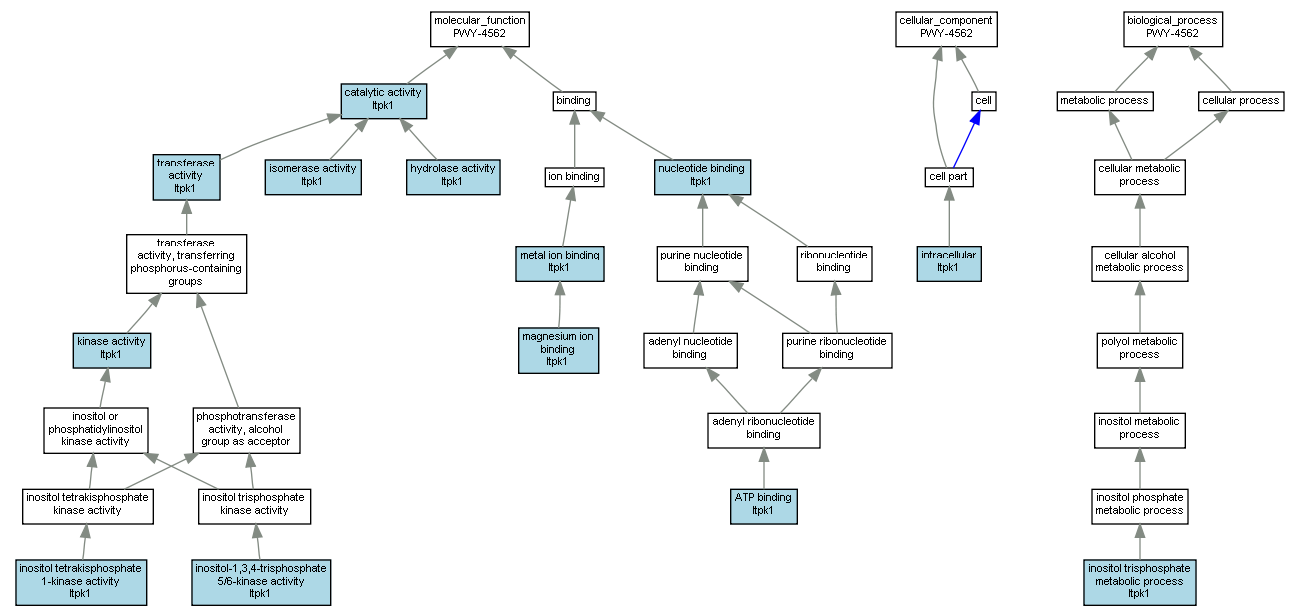
| Category | ID | Term | Mouse gene |
Evidence | Color Key |
| Molecular Function | GO:0005524 | ATP binding |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0003824 | catalytic activity |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0016787 | hydrolase activity |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0047325 | inositol tetrakisphosphate 1-kinase activity |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0035300 | inositol-1,3,4-trisphosphate 5/6-kinase activity |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0016853 | isomerase activity |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0016301 | kinase activity |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0000287 | magnesium ion binding |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0046872 | metal ion binding |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0000166 | nucleotide binding |
Itpk1 |
IEA | color key |
| Molecular Function | GO:0016740 | transferase activity |
Itpk1 |
IEA | color key |
| Cellular Component | GO:0005622 | intracellular |
Itpk1 |
IEA | color key |
| Biological Process | GO:0032957 | inositol trisphosphate metabolic process |
Itpk1 |
IEA | color key |