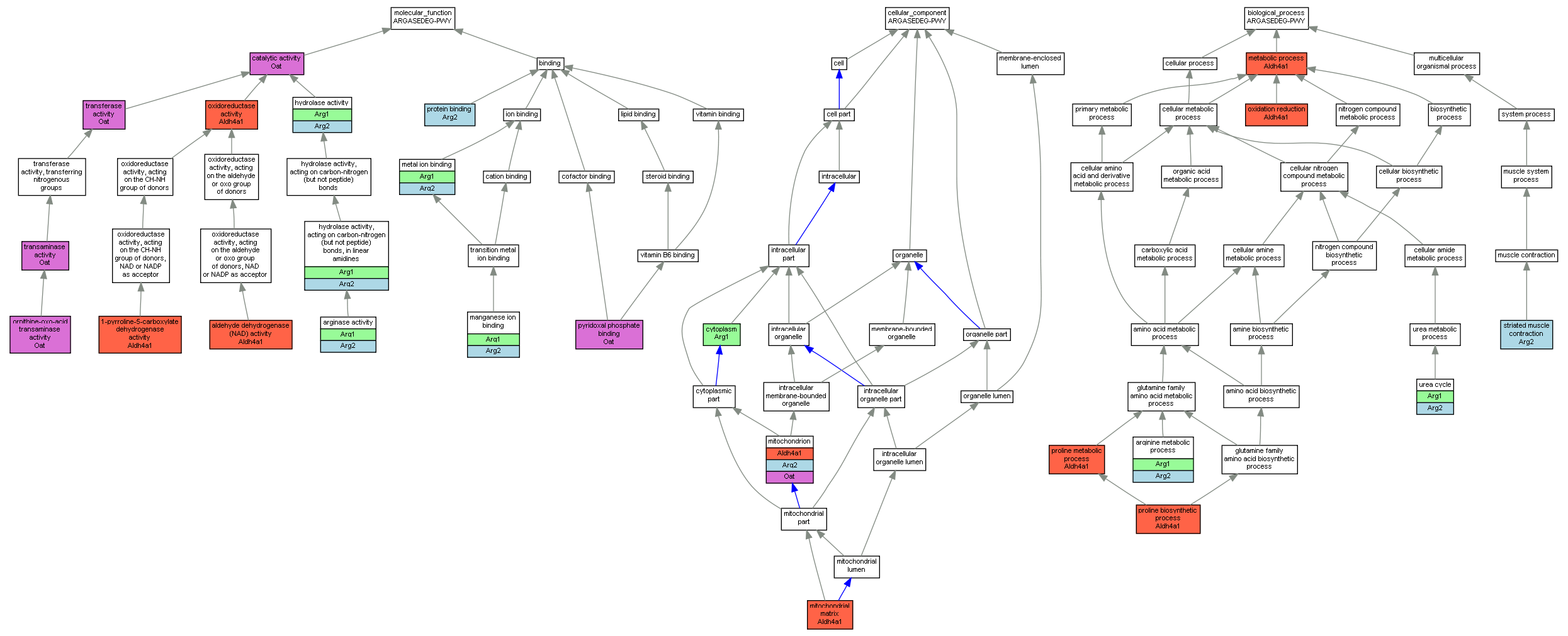
| Category | ID | Term | Mouse gene |
Evidence | Color Key |
| Molecular Function | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity |
Aldh4a1 |
IEA | color key |
| Molecular Function | GO:0004029 | aldehyde dehydrogenase (NAD) activity |
Aldh4a1 |
IEA | color key |
| Molecular Function | GO:0004053 | arginase activity |
Arg1 |
IDA | color key |
| Molecular Function | GO:0004053 | arginase activity |
Arg2 |
ISO | color key |
| Molecular Function | GO:0003824 | catalytic activity |
Oat |
IEA | color key |
| Molecular Function | GO:0016787 | hydrolase activity |
Arg1 |
IEA | color key |
| Molecular Function | GO:0016787 | hydrolase activity |
Arg2 |
IEA | color key |
| Molecular Function | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines |
Arg1 |
IEA | color key |
| Molecular Function | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines |
Arg2 |
IEA | color key |
| Molecular Function | GO:0030145 | manganese ion binding |
Arg1 |
IEA | color key |
| Molecular Function | GO:0030145 | manganese ion binding |
Arg2 |
IEA | color key |
| Molecular Function | GO:0046872 | metal ion binding |
Arg1 |
IEA | color key |
| Molecular Function | GO:0046872 | metal ion binding |
Arg2 |
IEA | color key |
| Molecular Function | GO:0004587 | ornithine-oxo-acid transaminase activity |
Oat |
IEA | color key |
| Molecular Function | GO:0016491 | oxidoreductase activity |
Aldh4a1 |
IEA | color key |
| Molecular Function | GO:0005515 | protein binding |
Arg2 |
ISO | color key |
| Molecular Function | GO:0030170 | pyridoxal phosphate binding |
Oat |
IEA | color key |
| Molecular Function | GO:0008483 | transaminase activity |
Oat |
IEA | color key |
| Molecular Function | GO:0016740 | transferase activity |
Oat |
IEA | color key |
| Cellular Component | GO:0005737 | cytoplasm |
Arg1 |
IEA | color key |
| Cellular Component | GO:0005759 | mitochondrial matrix |
Aldh4a1 |
IEA | color key |
| Cellular Component | GO:0005739 | mitochondrion |
Aldh4a1 |
IDA | color key |
| Cellular Component | GO:0005739 | mitochondrion |
Arg2 |
IDA | color key |
| Cellular Component | GO:0005739 | mitochondrion |
Arg2 |
ISO | color key |
| Cellular Component | GO:0005739 | mitochondrion |
Oat |
IDA | color key |
| Biological Process | GO:0006525 | arginine metabolic process |
Arg1 |
IEA | color key |
| Biological Process | GO:0006525 | arginine metabolic process |
Arg2 |
IEA | color key |
| Biological Process | GO:0008152 | metabolic process |
Aldh4a1 |
IEA | color key |
| Biological Process | GO:0055114 | oxidation reduction |
Aldh4a1 |
IEA | color key |
| Biological Process | GO:0006561 | proline biosynthetic process |
Aldh4a1 |
IEA | color key |
| Biological Process | GO:0006560 | proline metabolic process |
Aldh4a1 |
IEA | color key |
| Biological Process | GO:0006941 | striated muscle contraction |
Arg2 |
IMP | color key |
| Biological Process | GO:0000050 | urea cycle |
Arg1 |
IEA | color key |
| Biological Process | GO:0000050 | urea cycle |
Arg2 |
IEA | color key |